EXES Redux Developer’s Manual¶
Introduction¶
Document Purpose¶
This document is intended to provide all the information necessary to maintain the EXES Redux pipeline, used to produce Level 2 and 3 reduced products for EXES data, in either manual or automatic mode. Level 2 is defined as data that has been processed to correct for instrumental effects; Level 3 is defined as flux-calibrated data. A more general introduction to the data reduction procedure and the scientific justification of the algorithms is available in the EXES Redux User’s Manual.
This manual applies to EXES Redux version 3.0.0.
Redux Revision History¶
The EXES pipeline was originally developed as three separate packages: the EXES, which provided the image processing algorithms; FSpextool, which provided the spectral extraction algorithms and some supporting libraries; and Redux, which provided the interactive GUI, the automatic pipeline wrapper, and the supporting structure to call EXES and FSpextool algorithms.
EXES was a software package developed in IDL for the reduction of EXES data. Dr. John Lacy initially developed the algorithms in FORTRAN, for the reduction of data from TEXES (EXES’s predecessor instrument). The FORTRAN algorithms were translated into IDL and adapted for EXES by Melanie Clarke for the SOFIA DPS team in 2013 and 2014. Version 1.0.0 was released for use at SOFIA in 2015. The EXES IDL package was never used as a standalone data reduction package, so it did not provide interface scripts for its algorithms; it was designed specifically for incorporation into Redux.
FSpextool was built on top of a pre-release version of Spextool 4, an IDL-based package developed by Dr. Michael Cushing and Dr. William Vacca for the reduction of data from the SpeX instrument on the NASA Infrared Telescope Facility (IRTF). Spextool was originally released in October 2000, and has undergone a number of major and minor revisions since then. The last stable public release was v4.1, released January 2016. As Spextool does not natively support automatic command-line processing, FSpextool for SOFIA adapted the Spextool library to the SOFIA architecture and instruments; version 1.0.0 was originally released for use at SOFIA in July 2013.
Redux was developed to be a general-purpose interface to IDL data reduction algorithms. It provided an interactive GUI and an object-oriented structure for calling data reduction processes, but it does not provide its own data reduction algorithms. It was developed by Melanie Clarke for the SOFIA DPS team, to provide a consistent front-end to the data reduction pipelines for multiple instruments and modes, including EXES. It was first released in December 2013.
Between 2015 and 2021, the EXES Redux package was maintained by the EXES PI team, under Dr. Matthew Richter at UC Davis. The principal developers were Dr. Curtis DeWitt and Dr. Edward Montiel. In 2021, the SOFIA DPS team rebased the pipeline from the PI team version for release as EXES Redux v2.0.0, in support of EXES’s transition from PI class to facility class instrument.
In 2022, the EXES pipeline was entirely reimplemented in Python, as a set of software modules in the SOFIA Redux Python package:
sofia_redux.instruments.exes: processing algorithms specific to the EXES instrument
sofia_redux.spectroscopy: spectral extraction algorithms
sofia_redux.pipeline: interactive and batch mode interface tools for managing data reduction processes
sofia_redux.toolkit: numerical algorithms and supporting utilities
sofia_redux.visualization: data analysis and visualization tools
The exes module reimplements the algorithms in the EXES IDL package,
pipeline reimplements the Redux tools, and spectroscopy reimplements
FSpextool algorithms.
The SOFIA Redux package was developed as a unified Python package to support data reduction for all facility class instruments for SOFIA, replacing all legacy pipelines with an integrated, shared code base. The package was developed by the SOFIA DPS team, starting in 2018. The principal developers for SOFIA Redux prior to the EXES 3.0.0 release were Daniel Perera, Dr. Rachel Vander Vliet, and Melanie Clarke, for the SOFIA DPS team, with additional contributions from Dr. Karishma Bansal.
Overview of Software Structure¶
The sofia_redux package has several sub-modules organized by functionality:
sofia_redux
├── calibration
├── instruments
│ ├── exes
│ ├── fifi_ls
│ ├── flitecam
│ ├── forcast
│ └── hawc
├── pipeline
├── scan
├── spectroscopy
├── toolkit
└── visualization
The modules used in the EXES pipeline are described below.
sofia_redux.instruments.exes¶
The sofia_redux.instruments.exes package is written in Python using
standard scientific tools and libraries.
The data reduction algorithms used by the pipeline are straight-forward functions that generally take a data array, corresponding to a single image file, as an argument and return the processed image array as a result. They generally also take as secondary input a variance array to process alongside the image, a header structure to track metadata, and keyword parameters to specify non-default settings.
The exes module also stores any reference data needed by the EXES
pipeline. This includes bad pixel masks, nonlinearity coefficients,
dark files, and default header values and distortion correction parameters.
The default files may vary by date; these defaults are managed by the
readhdr function in the exes module. New date configurations may be added
to the caldefault.txt file in exes/data/caldefault.txt.
sofia_redux.toolkit¶
sofia_redux.toolkit is a repository for classes and functions of general
usefulness, intended to support multiple SOFIA pipelines. It contains several
submodules, for interpolation, image manipulation, multiprocessing support,
numerical calculations, and FITS handling. The utilities used by EXES are
generally simple functions that take input as arguments and return output
values.
sofia_redux.spectroscopy¶
The sofia_redux.spectroscopy package contains a library of general-purpose
spectroscopic functions. The EXES pipeline uses these algorithms
for spectroscopic image rectification, aperture identification, and
spectral extraction. Most of these algorithms are simple
functions that take spectroscopic data as input and return processed data
as output. However, the input and output values may be more complex than the
image processing algorithms in the exes package. The Redux interface
in the pipeline package manages the input and output requirements for
EXES data and calls each function individually. See the
sofia_redux.spectroscopy API documentation for more information.
sofia_redux.visualization¶
The sofia_redux.visualization package contains plotting and display
routines, relating to visualizing SOFIA data. For the EXES pipeline,
this package currently provides a module that supports generating
quick-look preview images.
sofia_redux.pipeline¶
Design¶
Redux is designed to be a light-weight interface to data reduction pipelines. It contains the definitions of how reduction algorithms should be called for any given instrument, mode, or pipeline, in either a command-line interface (CLI) or graphical user interface (GUI) mode, but it does not contain the reduction algorithms themselves.
Redux is organized around the principle that
any data reduction procedure can be accomplished by running a linear
sequence of data reduction steps. It relies on a Reduction class that
defines what these steps are and in which order they should be run
(the reduction “recipe”). Reductions have an associated Parameter
class that defines what parameters the steps may accept. Because
reduction classes share common interaction methods, they can be
instantiated and called from a completely generic front-end GUI,
which provides the capability to load in raw data files, and then:
set the parameters for a reduction step,
run the step on all input data,
display the results of the processing,
and repeat this process for every step in sequence to complete the
reduction on the loaded data. In order to choose the correct
reduction object for a given data set, the interface uses a Chooser
class, which reads header information from loaded input files and
uses it to decide which reduction object to instantiate and return.
The GUI is a PyQt application, based around the Application
class. Because the GUI operations are completely separate from the
reduction operations, the automatic pipeline script is simply a wrapper
around a reduction object: the Pipe class uses the Chooser to
instantiate the Reduction, then calls its reduce method, which calls
each reduction step in order and reports any output files generated.
Both the Application and Pipe classes inherit from a common Interface
class that holds reduction objects and defines the methods for
interacting with them. The Application class additionally may
start and update custom data viewers associated with the
data reduction; these should inherit from the Redux Viewer class.
All reduction classes inherit from the generic Reduction class,
which defines the common interface for all reductions: how parameters
are initialized and modified, how each step is called.
Each specific reduction class must then define
each data reduction step as a method that calls the appropriate
algorithm.
The reduction methods may contain any code necessary to accomplish the data reduction step. Typically, a reduction method will contain code to fetch the parameters for the method from the object’s associated Parameters class, then will call an external data reduction algorithm with appropriate parameter values, and store the results in the ‘input’ attribute to be available for the next processing step. If processing results in data that can be displayed, it should be placed in the ‘display_data’ attribute, in a format that can be recognized by the associated Viewers. The Redux GUI checks this attribute at the end of each data reduction step and displays the contents via the Viewer’s ‘display’ method.
Parameters for data reduction are stored as a list of ParameterSet
objects, one for each reduction step. Parameter sets contain the key,
value, data type, and widget type information for every parameter.
A Parameters class may generate these parameter sets by
defining a default dictionary that associates step names with parameter
lists that define these values. This dictionary may be defined directly
in the Parameters class, or may be read in from an external configuration
file or software package, as appropriate for the reduction.
EXES Redux¶
To interface to the EXES pipeline algorithms, Redux defines the
EXESReduction and EXESParameters classes. See Fig. 112
for a sketch of the Redux classes used by the EXES pipeline. The EXESReduction
class calls the sofia_redux.instruments.exes reduction functions, with
support from the sofia_redux.toolkit and sofia_redux.spectroscopy packages.
The EXESParameters class defines default parameter values for all reduction
steps.
The EXESReduction holds definitions for all reduction steps for the EXES pipeline:
Load Data: calls
sofia_redux.instruments.exes.readhdrCoadd Readouts: calls
sofia_redux.instruments.exes.readrawand optionallysofia_redux.instruments.exes.derasterizeorsofia_redux.instruments.exes.correct_row_gainsMake Flat: calls
sofia_redux.instruments.exes.makeflatandsofia_redux.instruments.exes.wavecalDespike: calls
sofia_redux.instruments.exes.despikeDebounce: calls
sofia_redux.instruments.exes.debounceSubtract Nods: calls
sofia_redux.instruments.exes.diff_arrand optionallysofia_redux.instruments.exes.cirrusFlat Correct: calls
sofia_redux.instruments.exes.calibrateClean Bad Pixels: calls
sofia_redux.instruments.exes.cleanUndistort: calls
sofia_redux.instruments.exes.tortCorrect Calibration: calls
sofia_redux.instruments.exes.makeflat.bnuCoadd Pairs: calls
sofia_redux.instruments.exes.coaddand optionallysofia_redux.instruments.exes.spatial_shiftorsofia_redux.instruments.exes.submeanMake Profiles: calls
sofia_redux.spectroscopy.mkspatprofandsofia_redux.spectroscopy.rectifyLocate Apertures: calls
sofia_redux.spectroscopy.findaperturesSet Apertures: calls
sofia_redux.spectroscopy.getaperturesandsofia_redux.spectroscopy.mkapmaskSubtract Background: calls
sofia_redux.spectroscopy.extspec.col_subbgExtract Spectra: calls
sofia_redux.spectroscopy.extspecandsofia_redux.instruments.exes.get_atranCombine Spectra: calls
sofia_redux.toolkit.image.combine_imagesRefine Wavecal: calls
sofia_redux.instruments.exes.wavecalMerge Orders: calls
sofia_redux.spectroscopy.mergespecandsofia_redux.toolkit.image.coaddMake Spectral Map: calls
sofia_redux.visualization.quicklook.make_spectral_plot
The recipe attribute for the reduction class specifies the above steps, in that order.
If an intermediate file is loaded, its product type is identified from the PRODTYPE keyword in its header, and the prodtype_map attribute is used to identify the next step in the recipe. This allows reductions to be picked up at any point, from a saved intermediate file. For more information on the scientific goals and methods used in each step, see the EXES Redux User’s Manual.
The EXESReduction class also contains several helper functions, that
assist in reading and writing files on disk, and identifying which
data to display in the interactive GUI. Display is performed via
the QADViewer class provided by the Redux package. Spatial profiles
and aperture locations are additionally displayed by the MatplotlibViewer
class. One-dimensional spectra are displayed by the EyeViewer class.
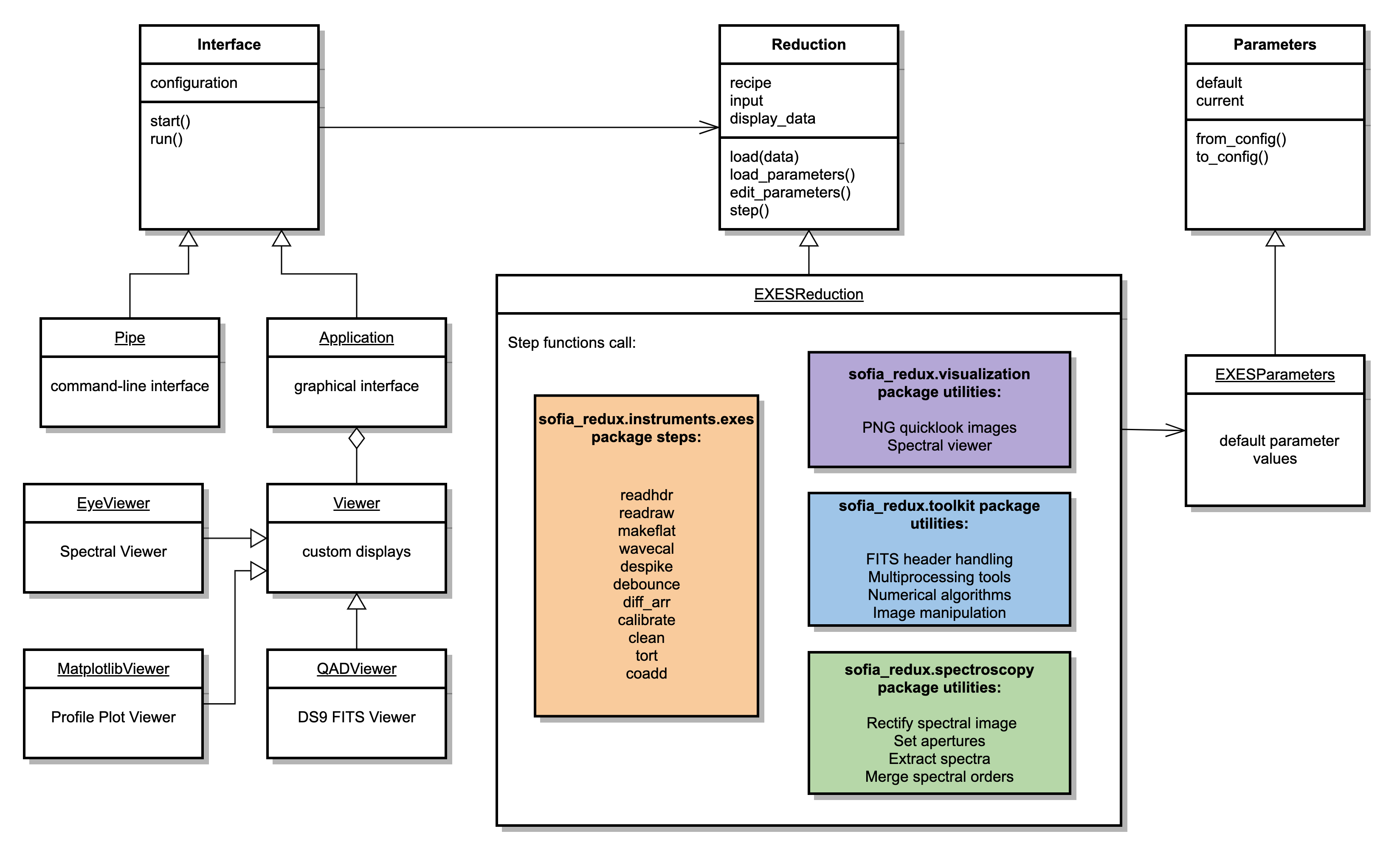
Fig. 112 EXES Redux class diagram.¶
Detailed Algorithm Information¶
The following sections list detailed information on the functions and procedures most likely to be of interest to the developer.
sofia_redux.instruments.exes¶
sofia_redux.instruments.exes.calibrate Module¶
Functions¶
|
Calibrate spectral image to physical units. |
sofia_redux.instruments.exes.cirrus Module¶
Functions¶
|
Correct nod-off-slit data for residual sky noise. |
sofia_redux.instruments.exes.clean Module¶
Functions¶
|
Correct bad pixels. |
sofia_redux.instruments.exes.coadd Module¶
Functions¶
|
Combine individual frames to increase signal-to-noise. |
sofia_redux.instruments.exes.correct_row_gains Module¶
Functions¶
|
Correct odd/even row gain offsets. |
sofia_redux.instruments.exes.debounce Module¶
Functions¶
|
Correct for optics shifts between nods (bounces). |
sofia_redux.instruments.exes.derasterize Module¶
Functions¶
|
Read and recombine a rasterized flat file. |
sofia_redux.instruments.exes.derive_tort Module¶
Functions¶
|
Derive distortion parameters and identify orders and illuminated regions. |
sofia_redux.instruments.exes.despike Module¶
Functions¶
|
Correct outlier pixels due to temporal spikes. |
sofia_redux.instruments.exes.diff_arr Module¶
Subtract sky frames or nod pairs.
Functions¶
|
Subtract sky frames, nod pairs, or a dark frame. |
sofia_redux.instruments.exes.get_atran Module¶
Functions¶
Clear all data from the atran cache. |
|
|
Retrieves atmospheric transmission data from the atran cache. |
|
Store atmospheric transmission data in the atran cache. |
|
Retrieve reference atmospheric transmission data. |
sofia_redux.instruments.exes.get_badpix Module¶
Functions¶
|
Get a bad pixel mask from a reference file. |
sofia_redux.instruments.exes.get_resolution Module¶
Functions¶
|
Retrieve expected spectral resolving power. |
sofia_redux.instruments.exes.lincor Module¶
Functions¶
|
Correct raw readout frames for detector nonlinearity. |
sofia_redux.instruments.exes.make_template Module¶
Functions¶
|
Make a spectral template for weighting and shifting spectra. |
sofia_redux.instruments.exes.makeflat Module¶
Functions¶
|
Generate calibrated flat frame; set distortion parameters. |
|
Black-body photon function. |
|
Black-body intensity function. |
|
Calibration factor for EXES blackbody source + flat mirror. |
sofia_redux.instruments.exes.mergehdr Module¶
Functions¶
|
Merge input headers. |
sofia_redux.instruments.exes.readhdr Module¶
Functions¶
|
Read and update an EXES FITS header. |
sofia_redux.instruments.exes.readraw Module¶
Functions¶
|
Correct for nonlinearity combine individual readouts. |
sofia_redux.instruments.exes.spatial_shift Module¶
Functions¶
|
Shift spectra for spatial alignment. |
sofia_redux.instruments.exes.submean Module¶
Functions¶
|
Subtract residual sky background from nod-on-slit data by removing the mean value at each wavelength. |
sofia_redux.instruments.exes.tort Module¶
Functions¶
|
Correct image for optical distortion. |
sofia_redux.instruments.exes.tortcoord Module¶
Functions¶
|
Calculate undistorted image coordinates. |
sofia_redux.instruments.exes.utils Module¶
Functions¶
|
Parse the DETSEC keyword in a header or from a string. |
|
Check the data dimensions and return number of frames. |
|
Check variance dimensions for expected shape. |
|
Get a reset dark image from a file on disk. |
|
Set the TOTTIME key in the header to the total elapsed time. |
|
Parse the central wavenumber from the input header. |
sofia_redux.instruments.exes.wavecal Module¶
Functions¶
|
Generate a wavelength calibration map from the grating equation. |
sofia_redux.toolkit¶
sofia_redux.toolkit.convolve.base Module¶
Classes¶
|
Convolution class allowing error propagation. |
Class Inheritance Diagram¶
digraph inheritance555cb6bce0 { bgcolor=transparent; rankdir=LR; size=""; "ConvolveBase" [URL="../../../api/sofia_redux.toolkit.convolve.base.ConvolveBase.html#sofia_redux.toolkit.convolve.base.ConvolveBase",fillcolor=white,fontname="Vera Sans, DejaVu Sans, Liberation Sans, Arial, Helvetica, sans",fontsize=10,height=0.25,margin=0.25,shape=box,style="setlinewidth(0.5),filled",target="_top",tooltip="Convolution class allowing error propagation."]; "Model" -> "ConvolveBase" [arrowsize=1.2,arrowtail=empty,dir=back,style="setlinewidth(0.5)"]; "Model" [URL="../../../api/sofia_redux.toolkit.utilities.base.Model.html#sofia_redux.toolkit.utilities.base.Model",fillcolor=white,fontname="Vera Sans, DejaVu Sans, Liberation Sans, Arial, Helvetica, sans",fontsize=10,height=0.25,margin=0.25,shape=box,style="setlinewidth(0.5),filled",target="_top",tooltip="Base model Class for fitting N-dimensional data"]; }sofia_redux.toolkit.convolve.kernel Module¶
Functions¶
|
Apply a kernel over multiple features |
|
Convolve an N-dimensional array with a user defined kernel or fixed box. |
|
Apply a least-squares (Savitzky-Golay) polynomial filter |
Classes¶
|
Generic convolution with a kernel |
|
Convolution with a box kernel (mean) |
|
Convolve using Savitzky-Golay filter |
Class Inheritance Diagram¶
digraph inheritance7459f88665 { bgcolor=transparent; rankdir=LR; size=""; "BoxConvolve" [URL="../../../api/sofia_redux.toolkit.convolve.kernel.BoxConvolve.html#sofia_redux.toolkit.convolve.kernel.BoxConvolve",fillcolor=white,fontname="Vera Sans, DejaVu Sans, Liberation Sans, Arial, Helvetica, sans",fontsize=10,height=0.25,margin=0.25,shape=box,style="setlinewidth(0.5),filled",target="_top",tooltip="Convolution with a box kernel (mean)"]; "KernelConvolve" -> "BoxConvolve" [arrowsize=1.2,arrowtail=empty,dir=back,style="setlinewidth(0.5)"]; "ConvolveBase" [URL="../../../api/sofia_redux.toolkit.convolve.base.ConvolveBase.html#sofia_redux.toolkit.convolve.base.ConvolveBase",fillcolor=white,fontname="Vera Sans, DejaVu Sans, Liberation Sans, Arial, Helvetica, sans",fontsize=10,height=0.25,margin=0.25,shape=box,style="setlinewidth(0.5),filled",target="_top",tooltip="Convolution class allowing error propagation."]; "Model" -> "ConvolveBase" [arrowsize=1.2,arrowtail=empty,dir=back,style="setlinewidth(0.5)"]; "KernelConvolve" [URL="../../../api/sofia_redux.toolkit.convolve.kernel.KernelConvolve.html#sofia_redux.toolkit.convolve.kernel.KernelConvolve",fillcolor=white,fontname="Vera Sans, DejaVu Sans, Liberation Sans, Arial, Helvetica, sans",fontsize=10,height=0.25,margin=0.25,shape=box,style="setlinewidth(0.5),filled",target="_top",tooltip="Generic convolution with a kernel"]; "ConvolveBase" -> "KernelConvolve" [arrowsize=1.2,arrowtail=empty,dir=back,style="setlinewidth(0.5)"]; "Model" [URL="../../../api/sofia_redux.toolkit.utilities.base.Model.html#sofia_redux.toolkit.utilities.base.Model",fillcolor=white,fontname="Vera Sans, DejaVu Sans, Liberation Sans, Arial, Helvetica, sans",fontsize=10,height=0.25,margin=0.25,shape=box,style="setlinewidth(0.5),filled",target="_top",tooltip="Base model Class for fitting N-dimensional data"]; "SavgolConvolve" [URL="../../../api/sofia_redux.toolkit.convolve.kernel.SavgolConvolve.html#sofia_redux.toolkit.convolve.kernel.SavgolConvolve",fillcolor=white,fontname="Vera Sans, DejaVu Sans, Liberation Sans, Arial, Helvetica, sans",fontsize=10,height=0.25,margin=0.25,shape=box,style="setlinewidth(0.5),filled",target="_top",tooltip="Convolve using Savitzky-Golay filter"]; "ConvolveBase" -> "SavgolConvolve" [arrowsize=1.2,arrowtail=empty,dir=back,style="setlinewidth(0.5)"]; }sofia_redux.toolkit.convolve.filter Module¶
Functions¶
|
Apply Savitzky-Golay filter to an array of arbitrary features |
|
Creates the correct windows for given order and samples |
|
Edge enhancement Sobel filter for n-dimensional images. |
sofia_redux.toolkit.fitting.fitpeaks1d Module¶
Functions¶
|
Simple convenience lookup to get width parameter name for various models. |
|
Creates the object fitting a model to data |
|
A simple wrapper to fit model to the data |
|
Convolve a model with a box (or another model) |
|
Create the |
|
Perform an initial search for peaks in the data |
|
Return a background model with initialized parameters |
|
Refine the initial fit and return a set of models |
|
Fit peaks (and optionally background) to a 1D set of data. |
|
Default data preparation for |
|
Default peak guess function for |
sofia_redux.toolkit.fitting.polynomial Module¶
Functions¶
|
Returns exponents for given polynomial orders in arbitrary dimensions. |
|
Create a system of linear equations to solve n-D polynomials |
|
Create a system of linear equations |
|
Linear equation solution by Gauss-Jordan elimination and matrix inversion |
|
Evalulate polynomial coefficients at x |
|
Evaluate a polynomial in multiple features |
|
Calculate the zeroth order polynomial coefficients and covariance |
|
Fit a polynomial to data samples using linear least-squares. |
|
Fit a polynomial to data samples using Gauss-Jordan elimination. |
|
Solve for polynomial coefficients using non-linear least squares fit |
|
Fits polynomial coefficients to N-dimensional data. |
|
|
|
|
|
|
|
Least squares polynomial fit to a surface |
|
Evaluate 2D polynomial coefficients |
|
Interpolate 2D data using polynomial regression (global) |
Classes¶
|
Fits and evaluates polynomials in N-dimensions. |
Class Inheritance Diagram¶
digraph inheritance812590e9a1 { bgcolor=transparent; rankdir=LR; size=""; "Model" [URL="../../../api/sofia_redux.toolkit.utilities.base.Model.html#sofia_redux.toolkit.utilities.base.Model",fillcolor=white,fontname="Vera Sans, DejaVu Sans, Liberation Sans, Arial, Helvetica, sans",fontsize=10,height=0.25,margin=0.25,shape=box,style="setlinewidth(0.5),filled",target="_top",tooltip="Base model Class for fitting N-dimensional data"]; "Polyfit" [URL="../../../api/sofia_redux.toolkit.fitting.polynomial.Polyfit.html#sofia_redux.toolkit.fitting.polynomial.Polyfit",fillcolor=white,fontname="Vera Sans, DejaVu Sans, Liberation Sans, Arial, Helvetica, sans",fontsize=10,height=0.25,margin=0.25,shape=box,style="setlinewidth(0.5),filled",target="_top",tooltip="Fits and evaluates polynomials in N-dimensions."]; "Model" -> "Polyfit" [arrowsize=1.2,arrowtail=empty,dir=back,style="setlinewidth(0.5)"]; }sofia_redux.toolkit.image.adjust Module¶
Functions¶
|
Shift an image by the specified amount. |
|
Rotate an image. |
|
Rebins an array to new shape |
|
Shifts an image by x and y offsets |
|
Replicates IDL rotate function |
|
Un-rotates an image using IDL style rotation types |
|
Return the pixel offset between an image and a reference |
|
Upsampled DFT by matrix multiplication. |
sofia_redux.toolkit.image.coadd Module¶
Functions¶
|
Coadd total intensity or spectral images. |
sofia_redux.toolkit.image.combine Module¶
Functions¶
|
Combine input image arrays. |
sofia_redux.toolkit.image.fill Module¶
Functions¶
|
|
|
|
|
Interpolates over image using a mask. |
|
Fills in NaN values in an image |
|
Clip a polygon to a square unit pixel |
|
Finds all pixels at least partially inside a specified polygon |
|
Uses the shoelace method to calculate area of a polygon |
|
Get pixel weights - depreciated by polyfillaa |
sofia_redux.toolkit.image.resize Module¶
Functions¶
|
Replacement for |
sofia_redux.toolkit.image.smooth Module¶
Functions¶
|
Quick and simple cubic polynomial fit to surface - no checks |
|
Returns the coefficients necessary for bicubic interpolation. |
|
|
|
Fit a plane to distribution of points. |
|
Fits a smooth surface to data using J. |
sofia_redux.toolkit.image.utilities Module¶
Functions¶
|
Convert from |
|
Allow translation of modes for scipy versions >= 1.6.0 |
|
Clip the array to the range of original values. |
|
A drop in replacement for |
sofia_redux.toolkit.image.warp Module¶
Functions¶
|
Warp data using transformation defined by two sets of coordinates |
|
Performs polynomial spatial warping |
|
Warp an image by mapping 2 coordinate sets with a polynomial transform. |
|
Check if a transform is homographic. |
|
Apply a metric transform to the supplied coordinates. |
|
Apply coefficients to polynomial terms. |
|
Estimate the polynomial transform for (x, y) coordinates. |
|
Apply the warping between two sets of coordinates to another. |
|
Warp the indices of an array with a given shape using a polynomial. |
|
Warp an n-dimensional image according to a given coordinate transform. |
Classes¶
|
Initialize a polynomial transform. |
Class Inheritance Diagram¶
digraph inheritance2c28666683 { bgcolor=transparent; rankdir=LR; size=""; "ABC" [URL="https://docs.python.org/3/library/abc.html#abc.ABC",fillcolor=white,fontname="Vera Sans, DejaVu Sans, Liberation Sans, Arial, Helvetica, sans",fontsize=10,height=0.25,margin=0.25,shape=box,style="setlinewidth(0.5),filled",target="_top",tooltip="Helper class that provides a standard way to create an ABC using"]; "PolynomialTransform" [URL="../../../api/sofia_redux.toolkit.image.warp.PolynomialTransform.html#sofia_redux.toolkit.image.warp.PolynomialTransform",fillcolor=white,fontname="Vera Sans, DejaVu Sans, Liberation Sans, Arial, Helvetica, sans",fontsize=10,height=0.25,margin=0.25,shape=box,style="setlinewidth(0.5),filled",target="_top"]; "ABC" -> "PolynomialTransform" [arrowsize=1.2,arrowtail=empty,dir=back,style="setlinewidth(0.5)"]; }sofia_redux.toolkit.interpolate.interpolate Module¶
Functions¶
|
Shift an equally spaced array of data values by an offset |
|
Interpolate values containing NaNs |
|
Perform cubic spline (tensioned) interpolation |
|
Perform a sinc interpolation on a data set |
|
Perform linear interpolation at a single point. |
|
Perform linear interpolation at a single point with error propagation. |
|
Perform linear interpolation of errors |
|
Propagate errors using Delaunay triangulation in N-dimensions |
|
Propagate errors using linear interpolation in N-dimensions |
|
Find the effective index of a function value in an ordered vector with NaN handling. |
|
Finds the effective index of a function value in an ordered array. |
Classes¶
|
Fast interpolation on a regular grid |
Class Inheritance Diagram¶
digraph inheritanceb8ee21f950 { bgcolor=transparent; rankdir=LR; size=""; "Interpolate" [URL="../../../api/sofia_redux.toolkit.interpolate.interpolate.Interpolate.html#sofia_redux.toolkit.interpolate.interpolate.Interpolate",fillcolor=white,fontname="Vera Sans, DejaVu Sans, Liberation Sans, Arial, Helvetica, sans",fontsize=10,height=0.25,margin=0.25,shape=box,style="setlinewidth(0.5),filled",target="_top",tooltip="Fast interpolation on a regular grid"]; }sofia_redux.toolkit.stats.stats Module¶
Functions¶
|
Determines the outliers in a distribution of data |
|
(Robustly) averages arrays along arbitrary axes. |
|
Combines a data set using median |
|
Computes statistics on a data set avoiding deviant points if requested |
|
Computes a mask derived from data Median Absolute Deviation (MAD). |
sofia_redux.toolkit.utilities.base Module¶
Classes¶
|
Base model Class for fitting N-dimensional data |
Class Inheritance Diagram¶
digraph inheritanced18b721993 { bgcolor=transparent; rankdir=LR; size=""; "Model" [URL="../../../api/sofia_redux.toolkit.utilities.base.Model.html#sofia_redux.toolkit.utilities.base.Model",fillcolor=white,fontname="Vera Sans, DejaVu Sans, Liberation Sans, Arial, Helvetica, sans",fontsize=10,height=0.25,margin=0.25,shape=box,style="setlinewidth(0.5),filled",target="_top",tooltip="Base model Class for fitting N-dimensional data"]; }sofia_redux.toolkit.utilities.fits Module¶
Functions¶
|
Insert or replace a keyword and value in the header |
|
Add HISTORY message to a FITS header before the pipeline. |
|
Make a function to add HISTORY messages to a header, prefixed with a string. |
|
Retrieve the data and header from a FITS file |
|
Returns the header of a FITS file |
|
Returns the data from a FITS file |
|
Convert a FITS header to an array of strings |
|
Convert an array of strings to a FITS header |
|
Returns the HDUList from a FITS file |
|
Write a HDULists to disk. |
|
Get a key value from a header. |
|
Context manager to temporarily set the log level. |
|
Order headers based on contents. |
|
Merge input headers. |
sofia_redux.toolkit.utilities.func Module¶
Functions¶
|
Check for 'truthy' values. |
|
Check for valid numbers. |
|
Returns list sorted in a human friendly manner |
|
Check if a file exists, and optionally if it has the correct permissions. |
|
Convert a header datestring to seconds |
|
Convert a string to an int or float. |
|
Returns a slice of an array in arbitrary dimension. |
|
Sets a value to a valid number type |
|
Gaussian model for curve_fit |
|
Broadcast an array to the desired shape. |
|
Recursively update a dictionary |
|
|
|
|
|
Taylor expansion generator for Polynomial exponents |
|
Convert a number of bytes to a string with correct suffix |
|
Remove any samples containing NaNs from sample points |
|
Return a byte array the same size as the input array. |
|
Generate a 2-D Julia fractal image |
|
Derive a mask to trim NaNs from an array |
|
Emulates the behaviour of np.nansum for NumPy versions <= 1.9.0. |
sofia_redux.toolkit.utilities.multiprocessing Module¶
Functions¶
|
Returns the maximum number of CPU cores available |
|
Return the actual number of cores to use for a given number of jobs. |
|
Return a valid number of jobs in the range 1 <= jobs <= max_cores. |
|
Process a series of tasks in serial, or in parallel using joblib. |
|
Pickle a object and save to the given filename. |
|
Unpickle a string argument if it is a file, and return the result. |
|
Pickle a list of objects to a temporary directory. |
|
Restore pickle files to objects in-place. |
Return whether the process is running in the main thread. |
|
|
Context manager to temporarily log messages for unique processes/threads |
|
Context manager to output log messages during multiprocessing. |
|
Remove all temporary logging files/directories and handle log records. |
|
Return the results of the function in multitask and save log records. |
|
Store the log records in a pickle file rather than emitting. |
|
Wrap a function for use with |
Classes¶
A log handler for multitask. |
Class Inheritance Diagram¶
digraph inheritance7a3b669de2 { bgcolor=transparent; rankdir=LR; size=""; "Filterer" [fillcolor=white,fontname="Vera Sans, DejaVu Sans, Liberation Sans, Arial, Helvetica, sans",fontsize=10,height=0.25,margin=0.25,shape=box,style="setlinewidth(0.5),filled",tooltip="A base class for loggers and handlers which allows them to share"]; "Handler" [URL="https://docs.python.org/3/library/logging.html#logging.Handler",fillcolor=white,fontname="Vera Sans, DejaVu Sans, Liberation Sans, Arial, Helvetica, sans",fontsize=10,height=0.25,margin=0.25,shape=box,style="setlinewidth(0.5),filled",target="_top",tooltip="Handler instances dispatch logging events to specific destinations."]; "Filterer" -> "Handler" [arrowsize=1.2,arrowtail=empty,dir=back,style="setlinewidth(0.5)"]; "MultitaskHandler" [URL="../../../api/sofia_redux.toolkit.utilities.multiprocessing.MultitaskHandler.html#sofia_redux.toolkit.utilities.multiprocessing.MultitaskHandler",fillcolor=white,fontname="Vera Sans, DejaVu Sans, Liberation Sans, Arial, Helvetica, sans",fontsize=10,height=0.25,margin=0.25,shape=box,style="setlinewidth(0.5),filled",target="_top",tooltip="A log handler for multitask."]; "Handler" -> "MultitaskHandler" [arrowsize=1.2,arrowtail=empty,dir=back,style="setlinewidth(0.5)"]; }sofia_redux.spectroscopy¶
sofia_redux.spectroscopy.binspec Module¶
Functions¶
|
Bin a spectrum between lmin and lmax with bins delta wide |
sofia_redux.spectroscopy.earthvelocity Module¶
Functions¶
|
Find the radial LSR velocity towards sky coordinates. |
|
Calculate the Cartesian velocity of the Sun. |
|
Provide velocities of the Earth towards a celestial position. |
sofia_redux.spectroscopy.extspec Module¶
Functions¶
|
Fit background to a single column. |
|
Extracts spectra from a rectified spectral image. |
sofia_redux.spectroscopy.findapertures Module¶
Functions¶
|
Determine the position of the aperture(s) in a spatial profile. |
sofia_redux.spectroscopy.fluxcal Module¶
Functions¶
|
Get pixel shift between flux and correction curve. |
|
Calibrate and telluric correct spectral flux. |
sofia_redux.spectroscopy.getapertures Module¶
Functions¶
|
Determine aperture radii for extraction. |
sofia_redux.spectroscopy.getspecscale Module¶
Functions¶
|
Determines the scale factors for a _stack of spectra |
sofia_redux.spectroscopy.mergespec Module¶
Functions¶
|
Combine two spectra into a single spectrum |
sofia_redux.spectroscopy.mkapmask Module¶
Functions¶
|
Constructs a 2D aperture mask. |
sofia_redux.spectroscopy.mkspatprof Module¶
Functions¶
|
Construct average spatial profiles. |
sofia_redux.spectroscopy.radvel Module¶
Functions¶
|
Calculate the expected extrinsic radial velocity wavelength shift. |
sofia_redux.spectroscopy.readflat Module¶
Functions¶
|
Reads a Spextool flat field FITS image |
sofia_redux.spectroscopy.readwavecal Module¶
Functions¶
|
Read a Spextool wavecal file |
sofia_redux.spectroscopy.rectify Module¶
Functions¶
|
Construct average spatial profiles over multiple orders |
sofia_redux.spectroscopy.rectifyorder Module¶
Functions¶
|
Given arrays of x and y coordinates, interpolate to defined grids |
|
Trim rows and columns from the edges of the coordinate arrays. |
|
Construct average spatial profiles for a single order |
|
Update a FITS header with spectral WCS information. |
|
Construct average spatial profiles for a single order |
sofia_redux.spectroscopy.smoothres Module¶
Functions¶
|
Smooth a data to a constant resolution |
sofia_redux.spectroscopy.tracespec Module¶
Functions¶
|
Trace spectral continua in a spatially/spectrally rectified image. |
sofia_redux.visualization¶
sofia_redux.visualization.quicklook Module¶
Functions¶
|
Generate a map image from a FITS file. |
|
Generate a plot of spectral data. |
sofia_redux.visualization.redux_viewer Module¶
Classes¶
Redux Viewer interface to the Eye of SOFIA. |
Class Inheritance Diagram¶
digraph inheritance28e1a7d94e { bgcolor=transparent; rankdir=LR; size=""; "EyeViewer" [URL="../../../api/sofia_redux.visualization.redux_viewer.EyeViewer.html#sofia_redux.visualization.redux_viewer.EyeViewer",fillcolor=white,fontname="Vera Sans, DejaVu Sans, Liberation Sans, Arial, Helvetica, sans",fontsize=10,height=0.25,margin=0.25,shape=box,style="setlinewidth(0.5),filled",target="_top",tooltip="Redux Viewer interface to the Eye of SOFIA."]; "Viewer" -> "EyeViewer" [arrowsize=1.2,arrowtail=empty,dir=back,style="setlinewidth(0.5)"]; "Viewer" [URL="../../../api/sofia_redux.pipeline.viewer.Viewer.html#sofia_redux.pipeline.viewer.Viewer",fillcolor=white,fontname="Vera Sans, DejaVu Sans, Liberation Sans, Arial, Helvetica, sans",fontsize=10,height=0.25,margin=0.25,shape=box,style="setlinewidth(0.5),filled",target="_top",tooltip="Parent class for Redux data viewers."]; }sofia_redux.visualization.controller Module¶
Standalone front-end for Eye of SOFIA display tool.
Functions¶
|
The Eye of SOFIA spectral viewer. |
|
Parse command line arguments. |
|
Check arguments for validity. |
sofia_redux.visualization.eye Module¶
Classes¶
|
Run the Eye of SOFIA. |
Class Inheritance Diagram¶
digraph inheritancebb54a4d2b2 { bgcolor=transparent; rankdir=LR; size=""; "Eye" [URL="../../../api/sofia_redux.visualization.eye.Eye.html#sofia_redux.visualization.eye.Eye",fillcolor=white,fontname="Vera Sans, DejaVu Sans, Liberation Sans, Arial, Helvetica, sans",fontsize=10,height=0.25,margin=0.25,shape=box,style="setlinewidth(0.5),filled",target="_top",tooltip="Run the Eye of SOFIA."]; }sofia_redux.pipeline¶
The Redux application programming interface (API), including the EXES
interface classes, are documented in the sofia_redux.pipeline package.
Appendix A: Pipeline Recipe¶
This JSON document is the black-box interface specification for the EXES Redux pipeline, as defined in the Pipetools-Pipeline ICD.
{
"inputmanifest" : "infiles.txt",
"outputmanifest" : "outfiles.txt",
"env" : {
"DPS_PYTHON": "$DPS_SHARE/share/anaconda3/envs/exes/bin"
},
"knobs" : {
"REDUX_CONFIG" : {
"desc" : "Redux parameter file containing custom configuration.",
"type" : "string",
"default": "None"
}
},
"command" : "$DPS_PYTHON/redux_pipe infiles.txt -c $DPS_REDUX_CONFIG"
}