FLITECAM Redux User’s Manual¶
Introduction¶
The SI Pipeline Users Manual (OP10) is intended for use by both SOFIA Science Center staff during routine data processing and analysis, and also as a reference for General Investigators (GIs) and archive users to understand how the data in which they are interested was processed. This manual is intended to provide all the needed information to execute the SI Level 2 Pipeline, flux calibrate the results, and assess the data quality of the resulting products. It will also provide a description of the algorithms used by the pipeline and both the final and intermediate data products.
A description of the current pipeline capabilities, testing results, known issues, and installation procedures are documented in the SI Pipeline Software Version Description Document (SVDD, SW06, DOCREF). The overall Verification and Validation (V&V) approach can be found in the Data Processing System V&V Plan (SV01-2232). Both documents can be obtained from the SOFIA document library in Windchill.
This manual applies to FLITECAM Redux version 2.0.0.
SI Observing Modes Supported¶
FLITECAM instrument information¶
The First Light Infrared Test Camera (FLITECAM) is an infrared camera, operating in the 1.0 - 5.5 \(\mu m\) range. It has a set of broadband filters for imaging and a set of grisms and order sorting filters for medium resolution spectroscopy.
The FLITECAM imaging mode provides seeing-limited images at 1 - 3 \(\mu m\) and diffraction-limited images at 3 - 5.5 \(\mu m\) (McLean, et al. 2006). The array (InSb ALADDIN III) size is 1024 x 1024 with a pixel scale of 0.47 arcseconds per pixel. This configuration results in a field of view (FOV) of approximately 8.0 arcminutes, but a circular stop and coma at the edges of the image restrict the usable FOV to 5.6 arcminutes (see Fig. 43).
FLITECAM has two filter wheels that contain a set of broadband imaging filters, a suite of broad order sorting filters for spectroscopy, and a few narrow-band imaging filters. Available broadband imaging filters for FLITECAM are J, H, and K. The narrow band filters are \(P_\alpha\), \(P_\alpha\) Continuum, \(3.0 \mu m\) ice, and \(3.30 \mu m\) PAH. In addition, FLITECAM offers several filters with limited support: \(H_{wide}\), \(K_{wide}\), \(K_{long}\), L, nbL, L’, M, nbM, and L+M. Detailed information about filter characteristics, saturation limits, sensitivities and observation planning can be found in the FLITECAM chapter of the SOFIA Observer’s Handbook.
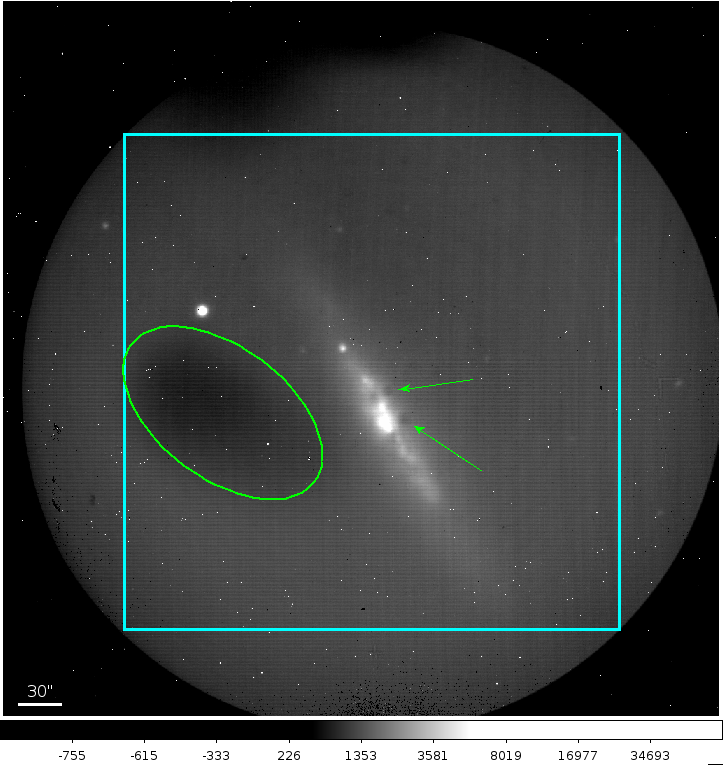
Fig. 43 A typical FLITECAM image obtained during a commissioning flight. The cyan box represents the usable portion of the array. Sources that fall outside of this box show too much coma for accurate PSF measurement. The small white dots visible in the image are hot pixels. The green ellipse encompasses a region of low quantum efficiency and the green arrows show obscurations in the optical path.¶
FLITECAM also has three grisms and five order-sorting filters that can be combined in different ways in order to access the full 1 - 5 \(\mu m\) wavelength range. It has two slits, a narrow slit (1 arcsecond) and a wide slit (~2 arcseconds), which allow for higher (R~2000) or lower (R~1300) resolution, respectively. The slits are both 60 arcseconds long and are adjacent to each other on the slit mask, such that spectra from both slits are acquired simultaneously during an integration (see Fig. 44).
FLITECAM was retired from SOFIA operations, effective February 2018. FLITECAM data is available for archival research via the SOFIA archive.

Fig. 44 A typical FLITECAM grism observation taken with the wide slit. As with the raw imaging data, there are hot pixels scattered across the frame. The wide-slit region is outlined in red; the narrow-slit region is visible to the right of the wide-slit region. Wavelength increases from the bottom to the top of the frame.¶
FLITECAM observing techniques¶
In any given pixel on a detector, the total number of counts is given by the sum of the counts from the dark current, telescope emission, sky background, and the target itself. Since the sky level can vary significantly over short timescales in the near infrared, it is typical to take pairs of observations back-to-back and subtract them in order to remove the dark current, telescope signal, and sky signal (to first order). An A frame exposure of the target is immediately followed by a B frame containing only sky at the target’s position:
Note that it is assumed that the sky level may vary somewhat between frame A and frame B, but the dark current and telescope emission do not. This residual sky emission is typically removed in the data reduction process.
For FLITECAM imaging, there are two observing modes: stare and nod-off-array. Both of these modes can be used with or without dithers. Most FLITECAM observations using are performed either in stare mode with dithers for uncrowded fields of view or nod-off-array with dithers for crowded and extended emission fields of view. In the first case, all dither positions are combined to produce a sky/flat frame to correct each image. In the second case, only the sky frames are combined to correct the on-source images.
FLITECAM grism observations offer two modes for producing A-B pairs. In the nod-along-slit mode, the A frame is taken with the target positioned one-third to one-quarter of the distance along the slit. After the A frame is complete, the telescope moves to place the target approximately the same distance from the other end of the slit. The exposure taken in this configuration is the B frame. It is typical, then, to repeat A-B pairs in either an A-B-A-B or A-B-B-A sequence until the total desired integration time is achieved. In this mode, the A frame provides the sky measurement at the target position for the B frame, and the B frame provides the sky for the A frame. This mode is useful as long as the target is compact, relative to the slit length.
In the second mode, nod-off-slit, the A frame is taken with the target at the center of the slit. The B frame is taken with the target completely off the slit, so that the exposure contains only the sky signal. In this mode, the B frame exists only to provide the sky measurement at the target position for the A frame, which may be useful if the target is significantly extended. In this mode, too, either the A-B-A-B or A-B-B-A observing sequence can be used.
Algorithm Description¶
Overview of data reduction steps¶
This section will describe, in general terms, the major algorithms that the FLITECAM Redux pipeline uses to reduce a FLITECAM observation.
The pipeline applies a number of corrections to each input file, regardless of the observing mode used to take the data. The initial steps used for imaging and grism modes are nearly identical. After preprocessing, individual images or spectra of a source must be combined to produce the final data product. This procedure depends strongly on the instrument configuration and observation mode.
See Fig. 45 and Fig. 46 for flowcharts of the processing steps used by the imaging and grism pipelines.
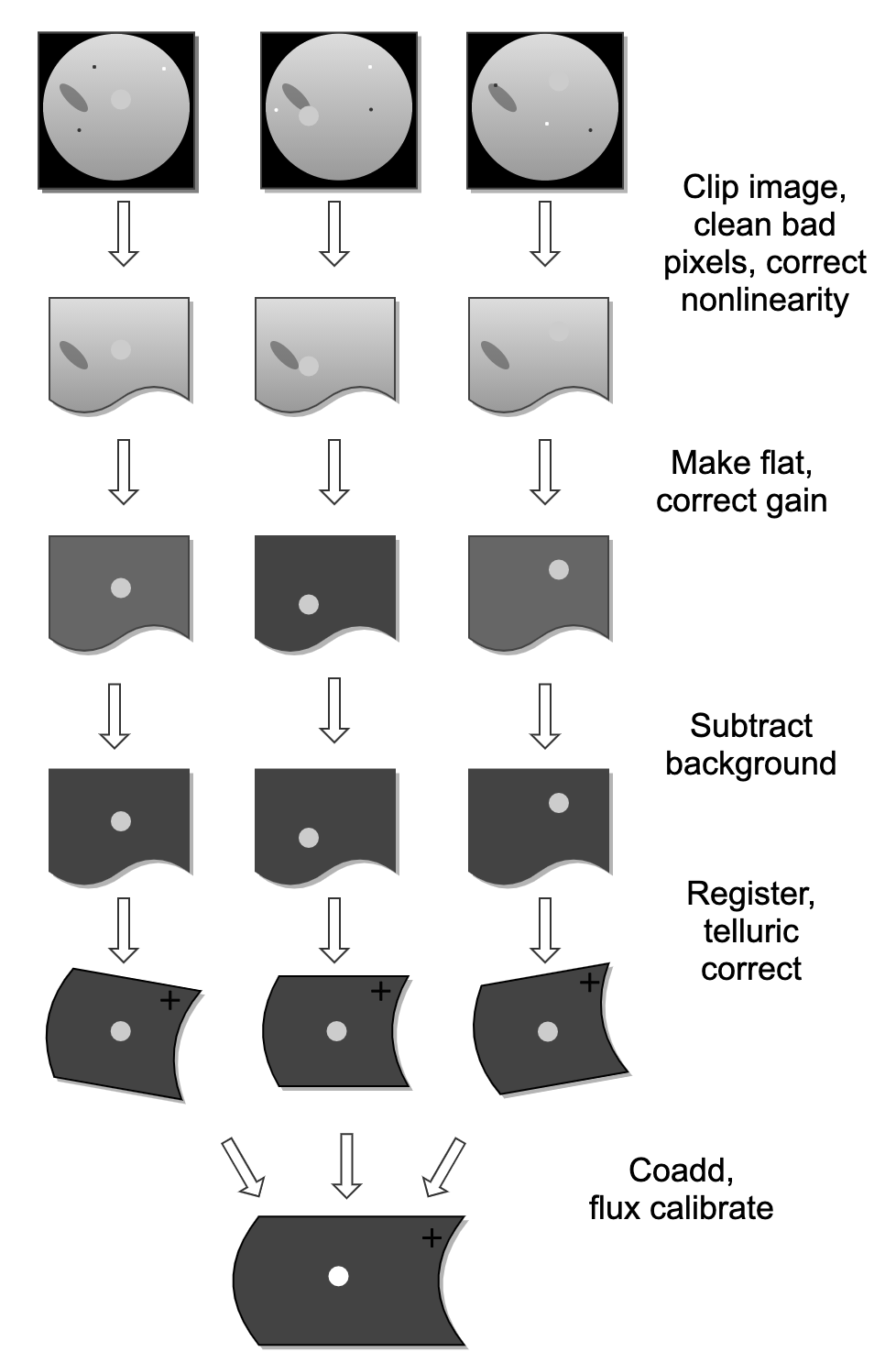
Fig. 45 Processing steps for imaging data.¶
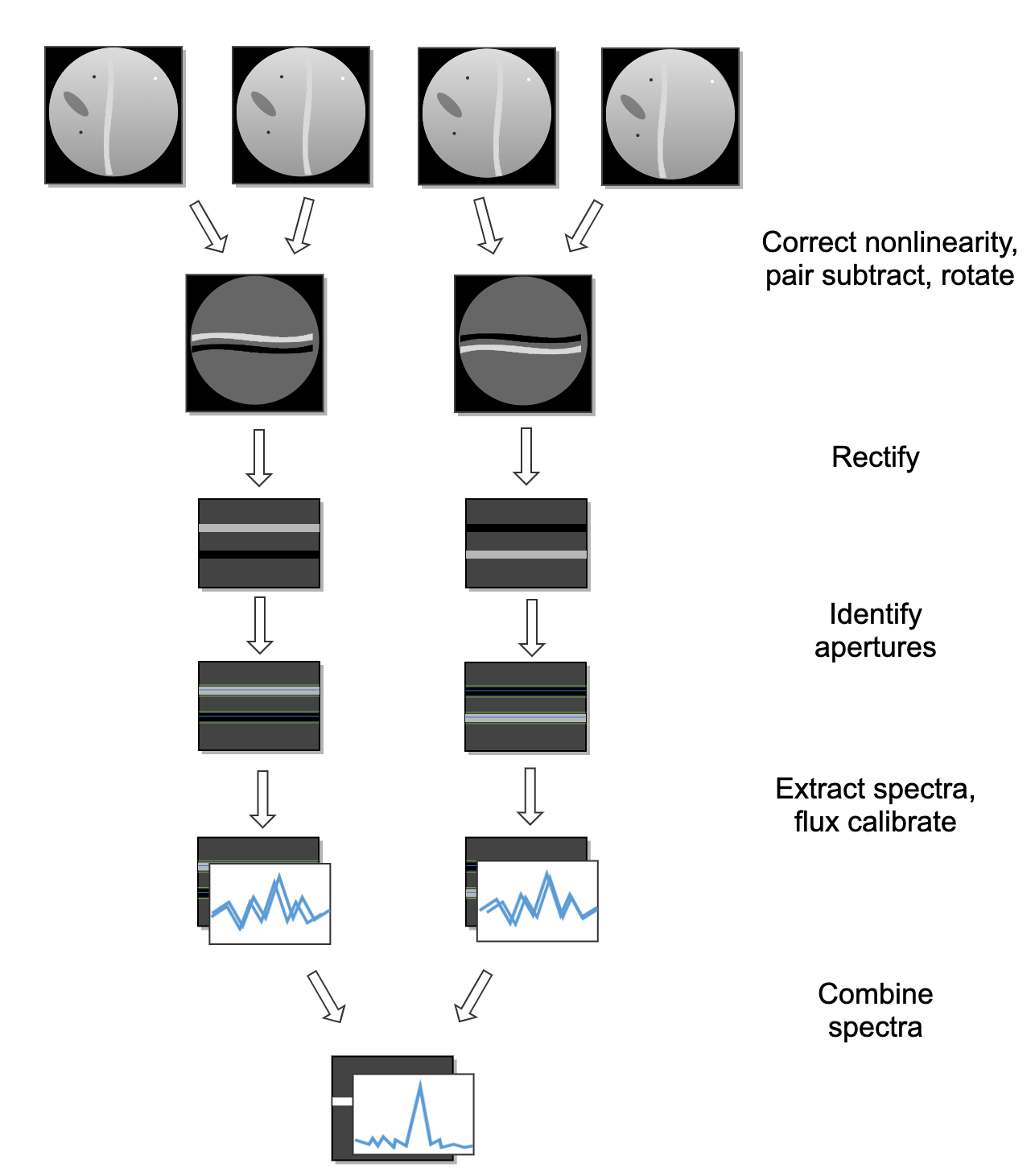
Fig. 46 Processing steps for grism data.¶
Reduction algorithms¶
The following subsections detail each of the data reduction pipeline steps:
Imaging steps
Correct for detector nonlinearity
Clip image and clean bad pixels
Correct gain
Subtract background
Register images
Correct for atmospheric transmission (telluric correct)
Coadd multiple observations
Calibrate flux
Spectroscopy steps
Correct for detector nonlinearity
Pair-subtract and rotate spectral images
Rectify spectral image
Identify apertures
Extract spectra
Calibrate flux and correct for atmospheric transmission
Combine multiple observations, or generate response spectra
Imaging steps¶
The following subsections detail each of the data reduction pipeline steps outlined in the imaging flowchart (Fig. 45).
Correct for detector nonlinearity¶
The first step of the imaging pipeline is to correct each input image for detector nonlinearity and generate an uncertainty image that associates an error value with each pixel in the flux image.
The nonlinearity coefficients for FLITECAM were determined by taking a series of flat exposures with varying exposure times. The count rates at each pixel in the flats were fit with a fourth order polynomial, and the resulting coefficients were stored in a FITS image file as a 3D data cube, where each plane corresponds to a different coefficient in the polynomial fit.
Following the Spextool nonlinearity paper (Vacca et al., 2004; see the Other Resources section), the coefficients are applied to the raw data as follows. First, the flat pedestal is determined from the first plane of the linearity coefficients:
where \(C_0\) is the first coefficient plane and \(\delta t_{flat}\) is the readout time for the flats used as input.
The pedestal for the image to be corrected is determined iteratively. The first estimate of the pedestal is:
where \(S_{tot}\) is the raw counts in the image, \(n_r\) is the number of readouts, \(n_c\) is the number of hardware coadds, \(\delta t\) is the readout time, \(\Delta t\) is the integration time, and f is a fractional value indicating how long it takes for an individual pixel to read out. Rigorously, f varies for each pixel, depending on its position in the array; for FLITECAM, an average value of f=0.5 is used for all pixels.
Using this pedestal estimate, the signal for an individual readout is estimated as:
and both pedestal and signal are corrected for nonlinearity. To account for the pedestal value of the flats used to determine the linearity coefficients, the coefficients are normalized by the first coefficient plane, and the polynomial is applied to the value to correct, minus the flat pedestal:
where \(C_{nl}\) is the full correction polynomial for each pixel in the image. This process is then repeated once more, replacing \(S_{tot}\) with
The final value for \(S_{tot}\), calculated from \(s^{(3)}\) and \(p^{(3)}\), is the linearity corrected image.
After linearity correction, the variance is calculated for each pixel from:
where g is the electronic gain and \(\sigma_r\) is the read noise for the detector (Vacca et al., 2004). This variance is propagated through all remaining reduction steps and its square root (the uncertainty) is recorded in all output files along with the image, as a separate extension in the file. [1]
Finally, the units in the flux and uncertainty images are converted from raw counts to counts per second by dividing by the integration time per coadd (\(\Delta t\)).
A bad pixel mask is also associated with the data after the nonlinearity correction step, in a BADMASK FITS extension. This initial mask marks any saturated pixels, recorded before linearity correction is applied, as bad pixels (0 = good, 1 = bad). These pixels are replaced with NaN values so that they do not propagate to subsequent steps.
Clip image and clean bad pixels¶
For the imaging pipeline, before proceeding, the linearity-corrected images and the corresponding uncertainty images are clipped to the size of the useful part of the detector (the cyan box in Fig. 43; see also Fig. 47).
Hot and cold bad pixels are then identified in the clipped image by iteratively searching for local outlier values. Bad pixels are replaced with NaN and their locations are added to the associated bad pixel mask.
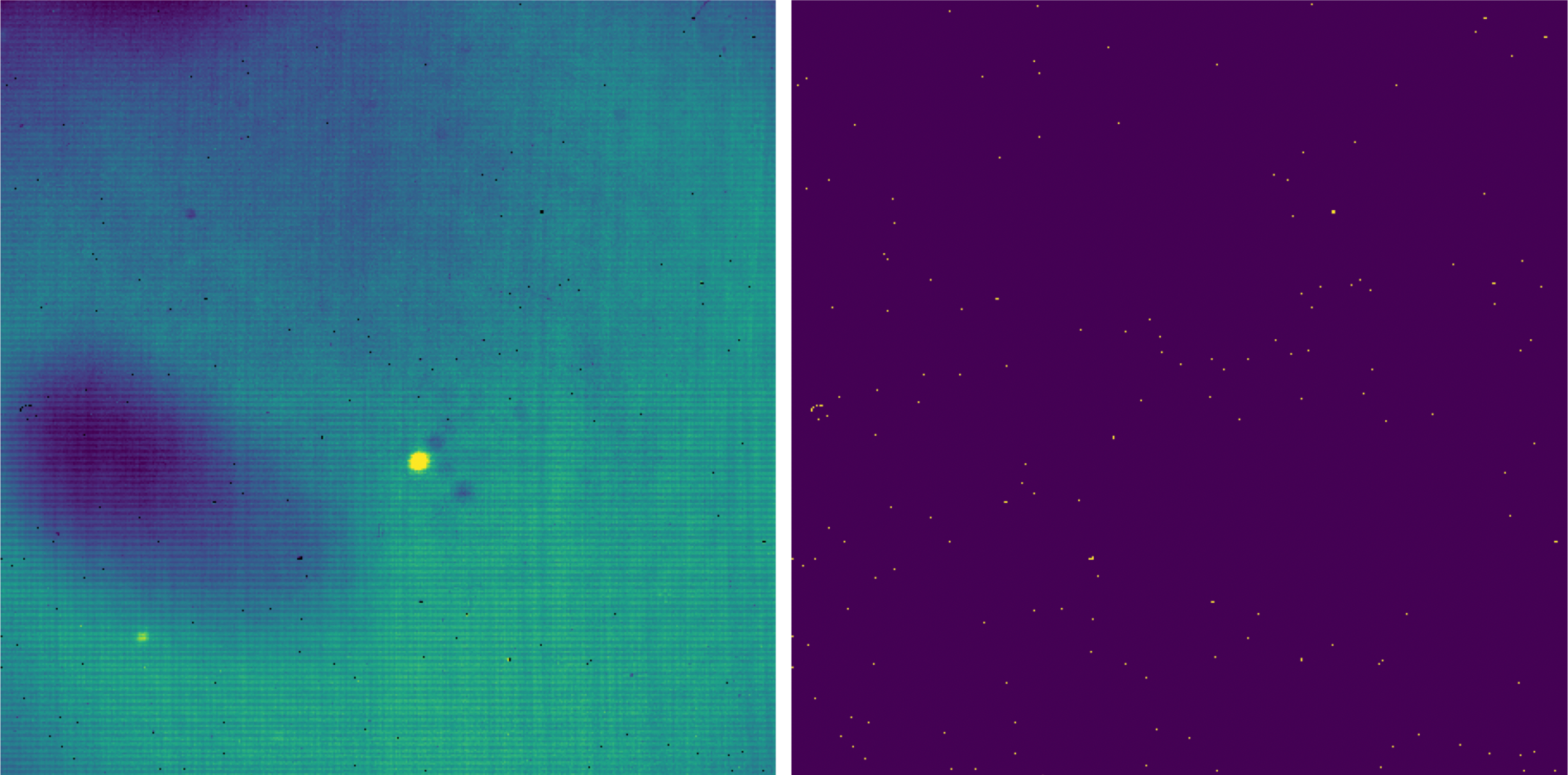
Fig. 47 Left: a clipped image, taken as part of a stare mode observation, corrected for nonlinearity and with bad pixels set to NaN. Right: the corresponding bad pixel mask.¶
Correct gain¶
As with all modern near-IR detectors, raw images produced by FLITECAM have significant pixel-to-pixel gain variations. In addition, FLITECAM’s detector has a large region of low quantum efficiency in the third quadrant and the top of the fourth quadrant of the detector, as shown in Fig. 43. These gain variations can be corrected by dividing by a normalized flat field image.
Imaging flats for FLITECAM are made from images of the sky. In nod-off-array mode, dithered sky images are used to generate a flat that is used to correct all on-source images. In stare mode, the dithered source images themselves are used to generate the flat. For each source image, a different flat is created from the remaining source images in order not to introduce correlations in the gain correction. In either case, the algorithm to generate the flat from the input files is the same.
First, all images are median-combined into a “draft” flat, with a sigma-clipping outlier rejection algorithm. The draft flat is used to flat-correct all input images. These draft images are then used to create object masks that identify any strong sources in the frame, via an image segmentation algorithm. The raw images are then scaled to the median value across all images and re-combined, ignoring any pixels identified in the object mask for each frame.
When the final flat frame has been created, it is divided by its median value to normalize it. This normalization value is stored in the FITS header in the FLATNORM keyword. This value may optionally be used later to correct for the sky background in the source images.
The final flat frame is divided into each source image to correct it for gain variations (Fig. 48).
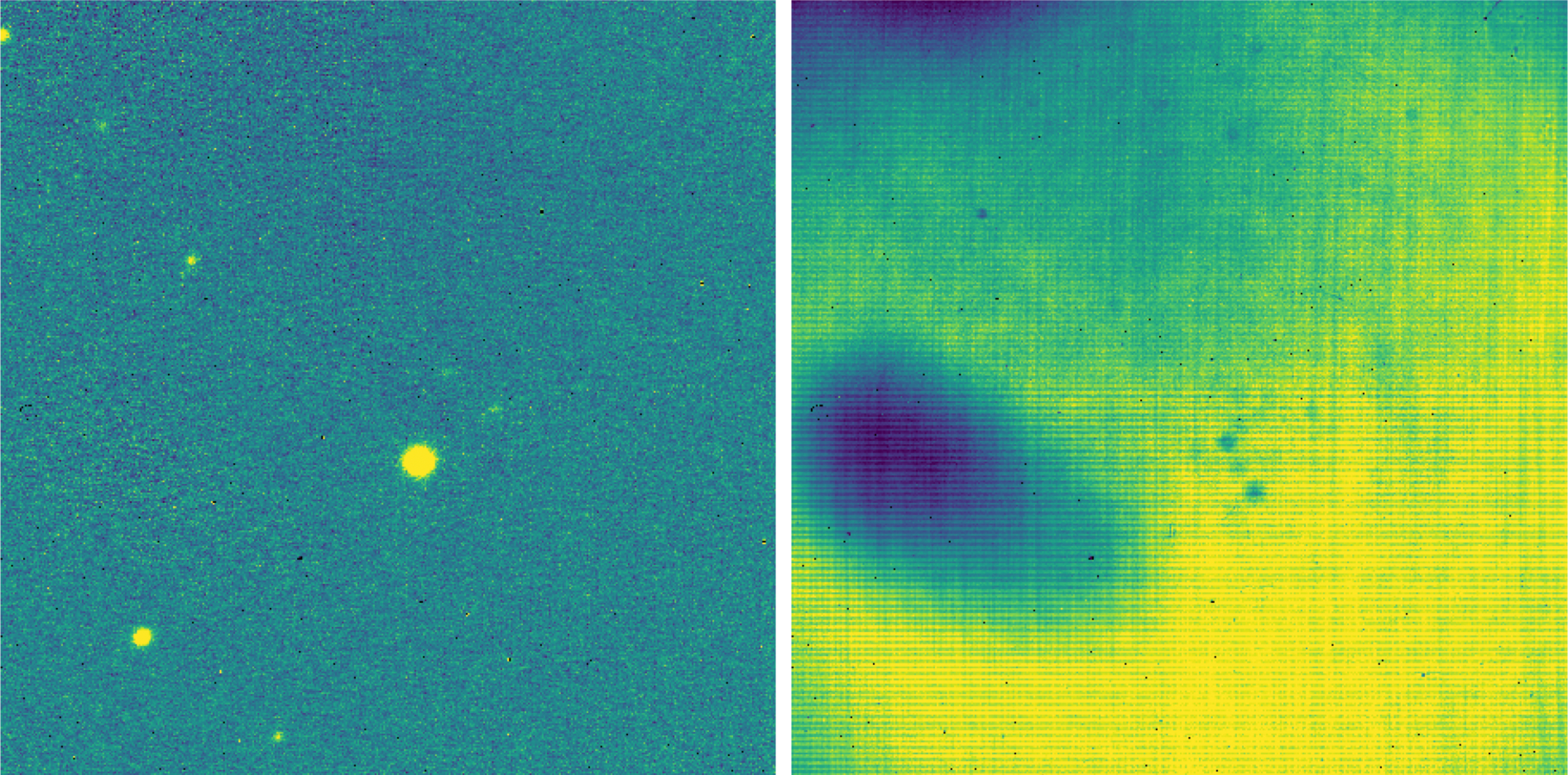
Fig. 48 Left: the stare mode image from Fig. 47, corrected for gain variations. Right: the normalized flat image used to correct the data, derived from the remaining dithers in the observation.¶
Subtract background¶
The sky background level must then be subtracted for each image. For imaging frames, since the flat is made from sky image, the average sky level is the median level of the unnormalized flat. This sky level is subtracted from each source image. Optionally, the median value of each individual image can be subtracted to correct for residual sky level variations, in place of the median level from the flat.
The sky level correction is recommended for observations of diffuse sources, for which emission fills the frame. The median level correction is recommended for observations in which the sky level varied significantly.
Register images¶
In order to combine multiple imaging observations of the same source, each image must be registered to a reference image, so that the pixels from each image correspond to the same location on the sky.
The registration information is typically encoded in the world coordinate system (WCS) embedded in each FITS file header. For most observations, the WCS is sufficiently accurate that no change is required in the registration step. However, if the WCS is faulty, it may be corrected in the registration step, using centroiding or cross-correlation between images to identify common sources. In this case,the first image is taken as the reference image, and calculated offsets are applied to the WCS header keywords (CRPIX1 and CRPIX2) in all subsequent images (Fig. 49). [2]
Earlier versions of this pipeline applied registration to the images themselves, rather than to the WCS in the FITS header, interpolating them into the same spatial grid. As of v2.0.0, registration affects only the CRPIX1 and CRPIX2 keywords in the header.
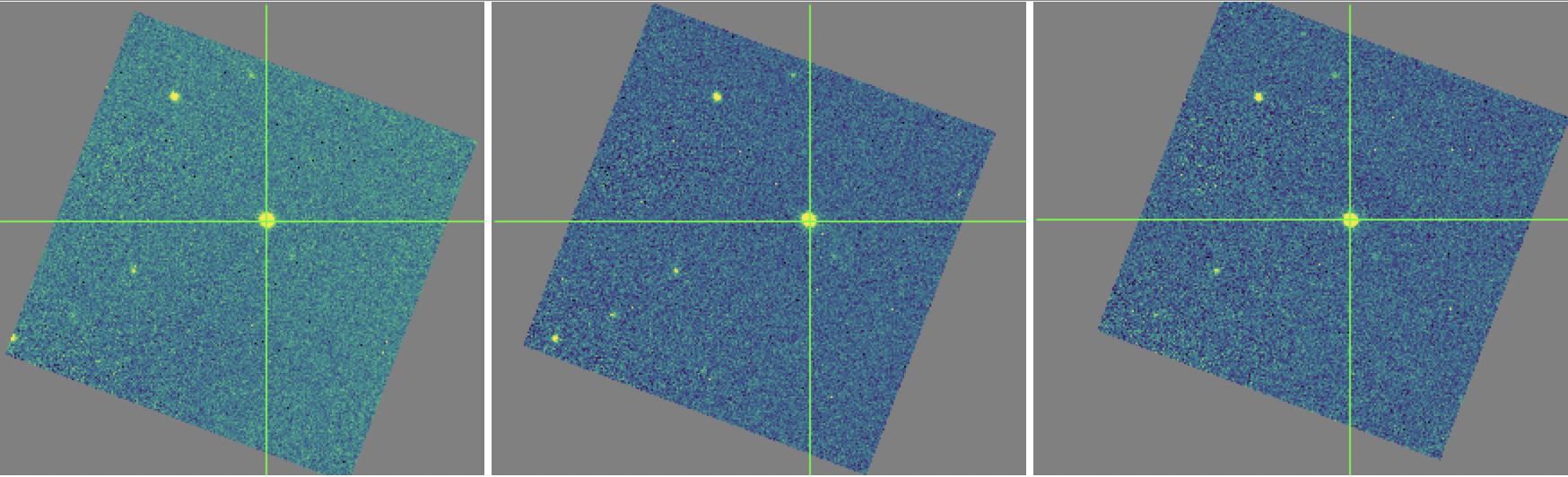
Fig. 49 Three dither positions from the stare mode observation of Fig. 47. The WCS was inaccurate for this observation, so the centroiding algorithm was used to correct the registration for these images. The registered images have not changed in dimension, but the FITS header keywords have been corrected to align them into a reference coordinate frame.¶
Correct for atmospheric transmission¶
For accurate flux calibration, the pipeline must first correct for the atmospheric opacity at the time of the observation. In order to combine images taken in different atmospheric conditions, or at different altitudes or zenith angles, the pipeline corrects the flux in each individual registered file for the estimated atmospheric transmission during the observations, based on the altitude and zenith angle at the time when the observations were obtained, relative to that computed for a reference altitude (41,000 feet) and reference zenith angle (45 degrees), for which the instrumental response has been calculated. The atmospheric transmission values are derived from the ATRAN code provided to the SOFIA program by Steve Lord. The pipeline applies the telluric correction factor directly to the flux in the image, and records it in the header keyword TELCORR.
After telluric correction, the pipeline performs aperture photometry on all observations that are marked as flux standards (FITS keyword OBSTYPE = STANDARD_FLUX). The brightest source in the field is fit with a Moffat profile to determine its centroid, and then its flux is measured, using an aperture of 12 pixels and a background region of 15-25 pixels. The aperture flux and error, as well as the fit characteristics, are recorded in the FITS header, to be used in the flux calibration process.
Coadd multiple observations¶
After registration and scaling, the pipeline coadds multiple observations of the same source with the same instrument configuration and observation mode. Each image is projected into the coordinate system of the first image, using its WCS to transform input coordinates into output coordinates. An additional offset may be applied for non-sidereal targets in order to correct for the motion of the target across the sky, provided that the target position is recorded in the FITS headers (TGTRA and TGTDEC). The projection is performed with a bilinear interpolation, then individual images are mean- or median-combined, with optional error weighting and robust outlier rejection.
For flux standards, photometry calculations are repeated on the coadded image, in the same way they were performed on the individual images.
Calibrate flux¶
For the imaging mode, flux calibration factors are typically calculated from all standards observed within a flight series. These calibration factors are applied directly to the flux images to produce an image calibrated to physical units. The final Level 3 product has image units of Jy per pixel (Fig. 50). [3]
See the flux calibration section, below, for more information.
Earlier versions of this pipeline did not produce a final calibrated file. Prior to v1.2.0, the final Level 3 products had image units of DN/sec, with the flux calibration factor (DN/sec/Jy) recorded in the FITS header keyword, CALFCTR. To convert these products to Jy/pixel, divide the flux image by the CALFCTR value.
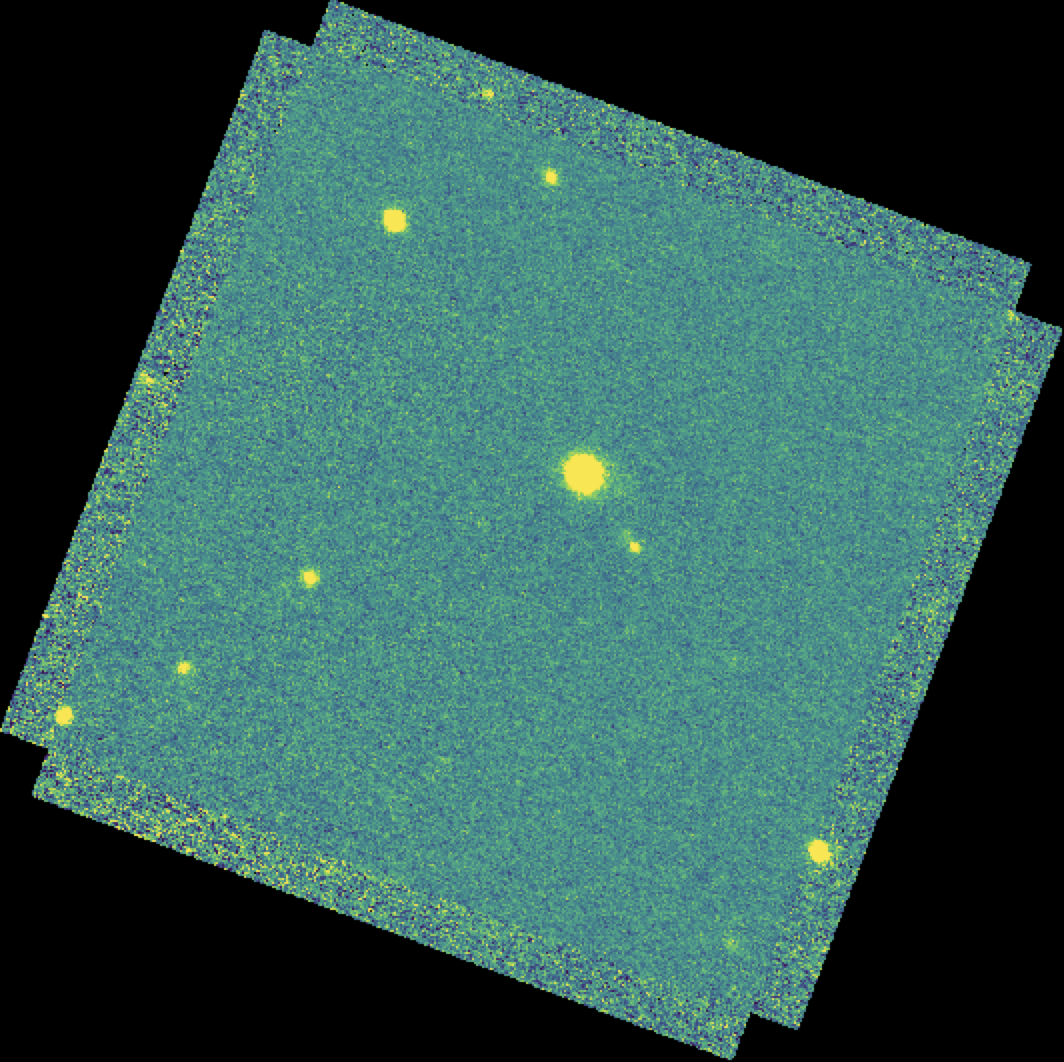
Fig. 50 The final coadded, calibrated image for the dithered stare mode observation of Fig. 47. The final image is rotated into a standard North-up, East-left orientation.¶
Mosaic¶
In some cases, it may be useful to stack together separate calibrated observations of the same target. In order to create a deeper image of a faint target, for example, observations taken across multiple flights may be combined together. Large maps may also be generated by taking separate observations, and stitching together the results. In these cases, the pipeline may register these files and coadd them, using the same methods as in the initial registration and coadd steps. The output product is a LEVEL_4 mosaic.
Spectroscopy Reduction algorithms¶
The following subsections detail each of the data reduction pipeline steps outlined in the grism flowchart (Fig. 46).
Image Processing¶
As for the FLITECAM imaging mode, the pipeline first corrects the input images for detector nonlinearity and creates an uncertainty image, using the algorithm described above, in the Correct for detector nonlinearity section). Then, the pipeline does A-B pair subtraction of all the input images. It also divides by a normalized flat image, if available. The resulting signal in the 2-D spectrum is:
where \(S_A\) is the corrected counts per second in frame A, \(S_B\) is the corrected counts per second in frame B, and flat is the normalized flat image.
Alongside the image processing, the individual variances for the A frame, B frame, and flat are combined as follows to get the total variance for the 2-D spectrum:
where \(V_A\) is the variance of frame A, \(V_B\) is the variance of frame B, and \(V_{flat}\) is the variance of the normalized flat image.
Stack common dithers¶
For very faint spectra, a stacking step may be optionally performed before spectral extraction. This step identifies spectra at common dither positions and mean- or median-combines them in order to increase signal-to-noise. This step may be applied if spectra are too faint to automatically identify appropriate apertures.
Rectify spectral image¶
For the spectroscopic mode, spatial and spectral distortions are corrected for by defining calibration images that assign a wavelength coordinate (in \(\mu m\)) and a spatial coordinate (in arcsec) to each detector pixel within the slit region of the detector (see Fig. 44). Each 2D spectral image in an observation is clipped and resampled into a rectified spatial-spectral grid, using these coordinates to define the output grid. If appropriate calibration data is available, the output from this step is an image in which wavelength values are constant along the columns, and spatial values are constant along the rows, correcting for any curvature in the spectral trace (Fig. 51).
The calibration maps used in rectification are generated from identifications of sky emission and telluric absorption lines and a polynomial fit to centroids of those features in pixel space for each row (i.e. along the dispersion direction). The derivation of a wavelength calibration is an interactive process, but application of the derived wavelength calibration is an automatic part of the data reduction pipeline. The default wavelength calibration is expected to be good to within approximately one pixel in the output spectrum.
For some observational cycles, sufficient calibration data may not be available, resulting in some residual spectral curvature, or minor wavelength calibration inaccuracies. The spectral curvature can be compensated for, in sources with strong continuum emission, by tracing the continuum center during spectral extraction (see next section). For other sources, a wider aperture may be set, at the cost of decreased signal-to-noise.
Additionally, a correction that accounts for spatial variations in the instrumental throughput may be applied to the rectified image. This “slit correction function” is a function of the position of the science target spectrum along the slit relative to that used for the standard stars. The slit function image is produced in a separate calibration process, from wavelength-rectified, averaged sky frames.
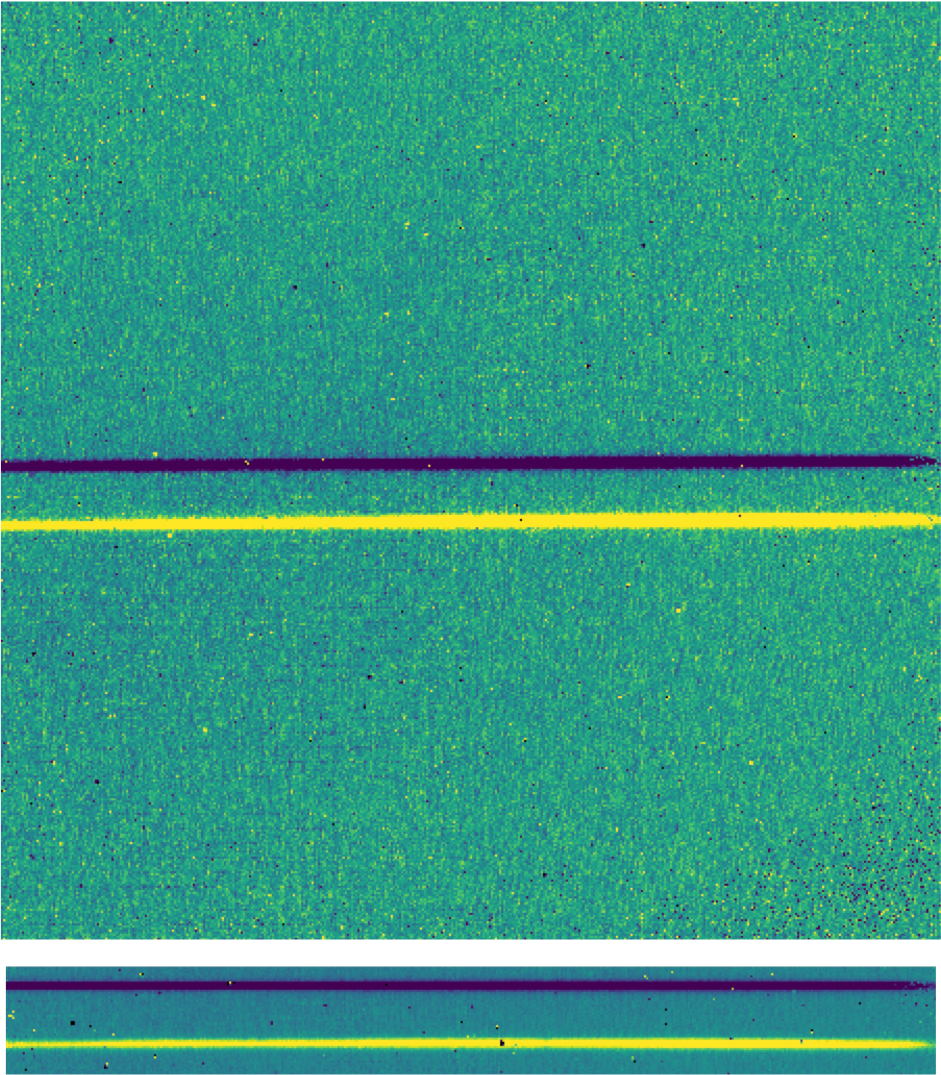
Fig. 51 A nod-along-slit spectral image after pair-subtraction, before (top) and after (bottom) rectification. Black spots indicate NaN values, marking saturated pixels identified during the nonlinearity correction step. Further bad pixels will be identified and ignored later in the extraction process.¶
Identify apertures¶
In order to aid in spectral extraction, the pipeline constructs a smoothed model of the relative intensity of the target spectrum at each spatial position, for each wavelength. This spatial profile is used to compute the weights in optimal extraction or to fix bad pixels in standard extraction (see next section). Also, the pipeline uses the median profile, collapsed along the wavelength axis, to define the extraction parameters.
To construct the spatial profile, the pipeline first subtracts the median signal from each column in the rectified spectral image to remove the residual background. The intensity in this image in column i and row j is given by
\(O_{ij} = f_{i}P_{ij}\)
where \(f_i\) is the total intensity of the spectrum at wavelength i, and \(P_{ij}\) is the spatial profile at column i and row j. To get the spatial profile \(P_{ij}\), we must approximate the intensity \(f_i\). To do so, the pipeline computes a median over the wavelength dimension (columns) of the order image to get a first-order approximation of the median spatial profile at each row \(P_j\). Assuming that
\(O_{ij} \approx c_{i}P_{j}\),
the pipeline uses a linear least-squares algorithm to fit \(P_j\) to \(O_{ij}\) and thereby determine the coefficients \(c_i\). These coefficients are then used as the first-order approximation to \(f_i\): the resampled order image \(O_{ij}\) is divided by \(f_i\) to derive \(P_{ij}\). The pipeline then fits a low-order polynomial along the columns at each spatial point s in order to smooth the profile and thereby increase its signal-to-noise. The coefficients of these fits can then be used to determine the value of \(P_{ij}\) at any column i and spatial point j (see Fig. 52, left). The median of \(P_{ij}\) along the wavelength axis generates the median spatial profile, \(P_j\) (see Fig. 52, right).
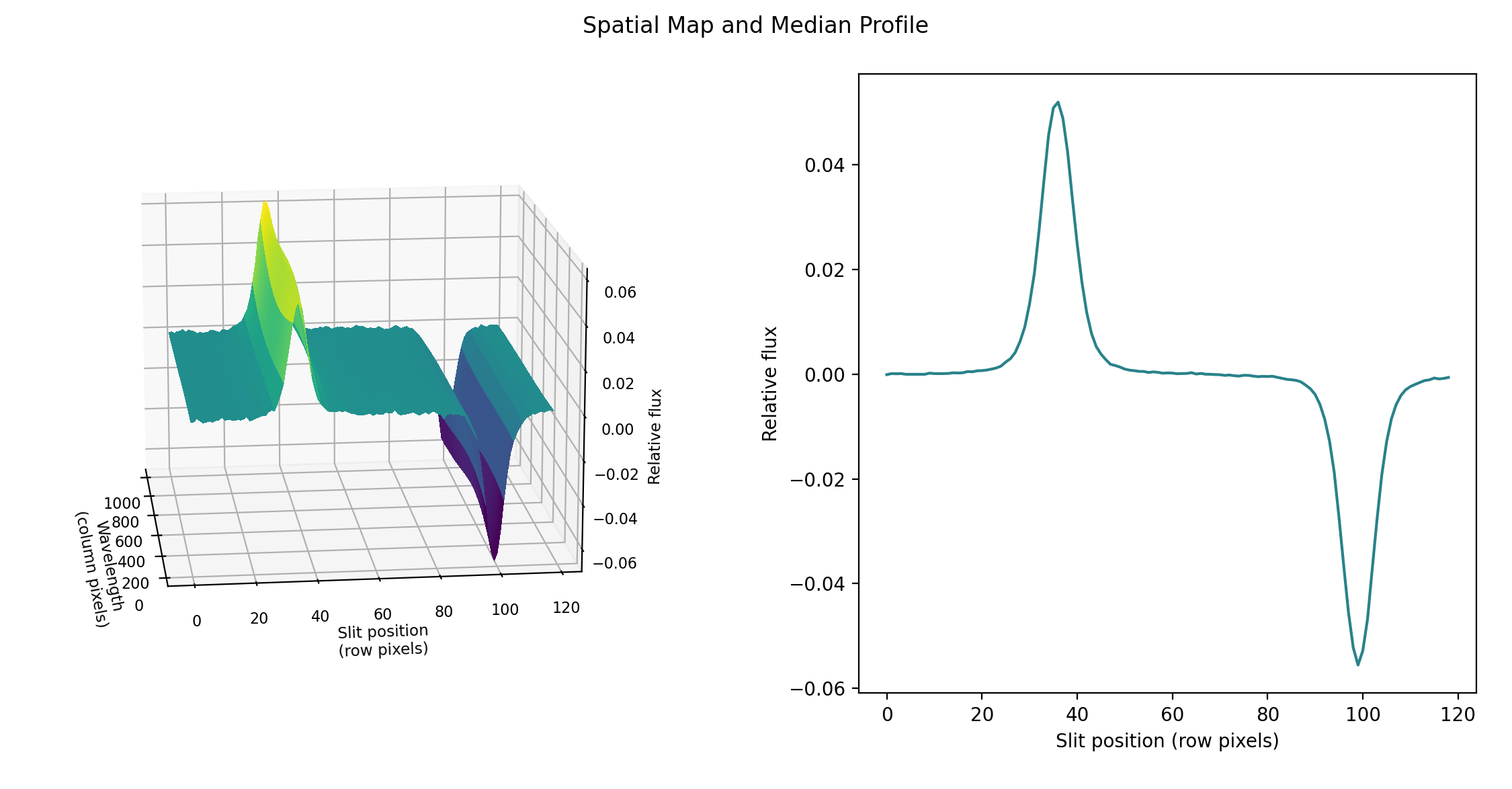
Fig. 52 Spatial model and median spatial profile, for the image in Fig. 51. The spatial model image here is rotated for comparison with the profile plot: the y-axis is along the bottom of the surface plot; the x-axis is along the left.¶
The pipeline then uses the median spatial profile to identify extraction apertures for the source. The aperture centers can be identified automatically by iteratively finding local maxima in the absolute value of the spatial profile, or can be specified directly by the user. By default, a single aperture is expected and defined for nod-off-slit mode; two apertures are expected for nod-along-slit mode.
The true position of the aperture center may vary somewhat with wavelength, as a result of small optical effects or atmospheric dispersion. To account for this variation, the pipeline attempts to trace the spectrum across the array. It fits a Gaussian in the spatial direction, centered at the specified position, at regular intervals in wavelength. The centers of these fits are themselves fitted with a low-order polynomial; the coefficients of these fits give the trace coefficients that identify the center of the spectral aperture at each wavelength. For extended sources, the continuum cannot generally be directly traced. Instead, the pipeline fixes the aperture center to a single spatial value.
Besides the aperture centers, the pipeline also specifies a PSF radius, corresponding to the distance from the center at which the flux from the source falls to zero. By default, this value is automatically determined from the width of a Gaussian fit to the peak in the median spatial profile, as
\(R_{psf} = 2.15 \cdot \text{FWHM}\).
For optimal extraction, the pipeline also identifies a smaller aperture radius, to be used as the integration region:
\(R_{ap} = 0.7 \cdot \text{FWHM}\).
This value should give close to optimal signal-to-noise for a Moffat or Gaussian profile. The pipeline also attempts to specify background regions outside of any extraction apertures, for fitting and removing the residual sky signal. All aperture parameters may be optionally overridden by the pipeline user.
Spectral extraction and merging¶
The spectral extraction algorithms used by the pipeline offer two different extraction methods, depending on the nature of the target source. For point sources, the pipeline uses an optimal extraction algorithm, described at length in the Spextool paper (see the Other Resources section, below, for a reference). For extended sources, the pipeline uses a standard summing extraction.
In either method, before extracting a spectrum, the pipeline first uses any identified background regions to find the residual sky background level. For each column in the 2D image, it fits a low-order polynomial to the values in the specified regions, as a function of slit position. This polynomial determines the wavelength-dependent sky level (\(B_{ij}\)) to be subtracted from the spectrum (\(D_{ij}\)).
The standard extraction method uses values from the spatial profile image (\(P_{ij}\)) to replace bad pixels and outliers, then sums the flux from all pixels contained within the PSF radius. The flux at column i is then:
\(f_{i,\text{sum}} = \sum_{j=j_1}^{j_2}(D_{ij} - B_{ij})\)
where \(j_1\) and \(j_2\) are the upper and lower limits of the extraction aperture (in pixels):
\(j_1 = t_i - R_{PSF}\)
\(j_2 = t_i + R_{PSF}\)
given the aperture trace center (\(t_i\)) at that column. This extraction method is the only algorithm available for extended sources.
Point sources may occasionally benefit from using standard extraction, but optimal extraction generally produces higher signal-to-noise ratios for these targets. This method works by weighting each pixel in the extraction aperture by how much of the target’s flux it contains. The pipeline first normalizes the spatial profile by the sum of the spatial profile within the PSF radius defined by the user:
\(P_{ij}^{'} = P_{ij} \Big/ \sum_{j=j_1}^{j_2}P_{ij}\).
\(P_{ij}^{'}\) now represents the fraction of the total flux from the target that is contained within pixel (i,j), so that \((D_{ij} - B_{ij}) / P_{ij}^{'}\) is a set of j independent estimates of the total flux at column i. The pipeline does a weighted average of these estimates, where the weight depends on the pixel’s variance and the normalized profile value. Then, the flux at column i is:
\(f_{i,\text{opt}} = \frac{\sum_{j=j_3}^{j_4}{M_{ij}P_{ij}^{'}(D_{ij} - B_{ij}) \big/ (V_{D_{ij}} + V_{B_{ij}})}}{\sum_{j=j_3}^{j_4}{M_{ij}{P_{ij}^{'}}^{2} \big/ (V_{D_{ij}} + V_{B_{ij}})}}\)
where \(M_{ij}\) is a bad pixel mask and \(j_3\) and \(j_4\) are the upper and lower limits given by the aperture radius:
\(j_3 = t_i - R_{ap}\)
\(j_4 = t_i + R_{ap}\)
Note that bad pixels are simply ignored, and outliers will have little effect on the average because of the weighting scheme.
The variance for the standard spectroscopic extraction is a simple sum of the variances in each pixel within the aperture. For the optimal extraction algorithm, the variance on the ith pixel in the extracted spectrum is calculated as:
where \(P_{ij}^{'}\) is the scaled spatial profile, \(M_{ij}\) is a bad pixel mask, \(V_{ij}\) is the variance at each background-subtracted pixel, and the sum is over all spatial pixels \(j\) within the aperture radius. The error spectrum for 1D spectra is the square root of the variance.
Calibrate flux and correct for atmospheric transmission¶
Extracted spectra are corrected individually for instrumental response and atmospheric transmission, a process that yields a flux-calibrated spectrum in units of Jy per pixel. See the section on flux calibration, below, for more detailed information.
The rectified spectral images are also corrected for atmospheric transmission, and calibrated to physical units in the same manner. Each row of the image is divided by the same correction as the 1D extracted spectrum. This image is suitable for custom extractions of extended fields: a sum over any number of rows in the image produces a flux-calibrated spectrum of that region, in the same units as the spectrum produced directly by the pipeline.
Note that the FITS header for the primary extension for this product (PRODTYPE = ‘calibrated_spectrum’) contains a full spatial and spectral WCS that can be used to identify the coordinates of any spectra so extracted. The primary WCS identifies the spatial direction as arcseconds up the slit, but a secondary WCS with key = ‘A’ identifies the RA, Dec, and wavelength of every pixel in the image. [4] Either can be extracted and used for pixel identification with standard WCS manipulation packages, such as the astropy WCS package.
After telluric correction, it is possible to apply a correction to the calibrated wavelengths for the motion of the Earth relative to the solar system barycenter at the time of the observation. For FLITECAM resolutions, we expect this wavelength shift to be a small fraction of a pixel, well within the wavelength calibration error, so we do not directly apply it to the data. The shift (as \(d\lambda / \lambda\)) is calculated and stored in the header in the BARYSHFT keyword. An additional wavelength correction to the local standard of rest (LSR) from the barycentric velocity is also stored in the header, in the LSRSHFT keyword.
The overall accuracy of the FLITECAM WCS in spectroscopic mode has not been studied in detail. In particular, for nod-along-slit data, no attempt has been made to co-align positive and negative spectral traces in the 2D spectral image. The reference position for the WCS may refer to one or neither of the spectral positions in the rectified 2D image.
Combine multiple observations¶
The final pipeline step for most grism observation modes is coaddition of multiple spectra of the same source with the same instrument configuration and observation mode. The individual extracted 1D spectra are combined with a robust weighted mean, by default. The 2D spectral images are also coadded, using the same algorithm as for imaging coaddition, and the spatial/spectral WCS to project the data into a common coordinate system.
Reductions of flux standards have an alternate final product (see Response spectra, below).
Response spectra¶
The final product of pipeline processing of telluric standards is not a calibrated, combined spectrum, but rather an instrumental response spectrum that may be used to calibrate science target spectra. These response spectra are generated from individual observations of calibration sources by dividing the observed spectra by a model of the source multiplied by an atmospheric model. The resulting response curves may then be combined with other response spectra from a flight series to generate a final instrument response spectrum that is used in calibrating science spectra. See the flux calibration section, below, for more information.
Other Resources¶
For more information on the instrument itself, see the FLITECAM paper:
FLITECAM: a 1-5 micron camera and spectrometer for SOFIA, Ian S. McLean, et al. (2006, SPIE 6269E, 168).
For more information on the algorithms used in spectroscopic data reduction, see the Spextool papers:
Spextool: A Spectral Extraction Package for SpeX, a 0.8-5.5 Micron Cross-Dispersed Spectrograph, Michael C. Cushing, William D. Vacca and John T. Rayner (2004, PASP 116, 362).
A Method of Correcting Near-Infrared Spectra for Telluric Absorption, William D. Vacca, Michael C. Cushing and John T. Rayner (2003, PASP 115, 389).
Nonlinearity Corrections and Statistical Uncertainties Associated with Near-Infrared Arrays, William D. Vacca, Michael C. Cushing and John T. Rayner (2004, PASP 116, 352).
Flux calibration¶
Imaging Flux Calibration¶
The reduction process, up through image coaddition, generates Level 2 images with data values in units of counts per second (ct/s). After Level 2 imaging products are generated, the pipeline derives the flux calibration factors (in units of ct/s/Jy) and applies them to each image. The calibration factors are derived for each FLITECAM filter configuration from observations of calibrator stars.
After the calibration factors have been derived, the coadded flux is divided by the appropriate factor to produce the Level 3 calibrated data file, with flux in units of Jy/pixel. The value used is stored in the FITS keyword CALFCTR.
Reduction steps¶
The calibration is carried out in several steps. The first step consists of measuring the photometry of all the standard stars for a specific mission or flight series, after the images have been corrected for the atmospheric transmission relative to that for a reference altitude and zenith angle [5]. The pipeline performs aperture photometry on the reduced Level 2 images of the standard stars after the registration stage using a photometric aperture radius of 12 pixels. The telluric-corrected photometry of the standard star is related to the measured photometry of the star via
where the ratio \(R_{\lambda}^{ref} / R_{\lambda}^{std}\) accounts for differences in system response (atmospheric transmission) between the actual observations and those for the reference altitude of 41000 feet and a telescope elevation of 45\(^\circ\). Similarly, for the science target, we have
Calibration factors (in ct/s/Jy) for each filter are then derived from the measured photometry (in ct/s) and the known fluxes of the standards (in Jy) in each filter. These predicted fluxes were computed by multiplying a model stellar spectrum by the overall filter + instrument + telescope + atmosphere (at the reference altitude and zenith angle) response curve and integrating over the filter passband to compute the mean flux in the band. The adopted filter throughput curves are those provided by the vendor. The instrument throughput is calculated by multiplying an estimate of the instrumental optics transmission(0.80) and the detector quantum efficiency (0.56). The FLITECAM overall throughput is (0.285). The telescope throughput value is assumed to be constant (0.85) across the entire FLITECAM wavelength range.
Photometric standards for FLITECAM have been chosen from three sources: (1) bright stars with spectral classifications of A0V as listed in SIMBAD; (2) Landolt SA stars (K giants and A0-4 main sequence stars) listed as ‘supertemplate’ stars in Cohen et al. (2003); K giant stars listed as ‘spectral template’ stars in Cohen et al. (1999). For all of these objects, models are either available (from the Cohen papers) or derivable (from a model of Vega for the A0V stars). Use of the A0V stars requires scaling the Vega model to the observed magnitudes of the target and reddening the model to match the observed color excess of the target. It should be noted that A0V stars should be used to calibrate the Pa alpha filter, as these objects have a strong absorption feature in this band. The models of the spectral template K giants listed in Cohen et al. (1999) extend down only to 1.2 microns, and therefore cannot be used to calibrate the J band filter.
The calibration factor, C, is computed from
with an uncertainty given by
Here, \(\lambda_{piv}\) is the pivot wavelength of the filter, and \(\langle \lambda \rangle\) is the mean wavelength of the filter. The calibration factor refers to a nominal flat spectrum source at the reference wavelength \(\lambda_{ref}\).
The calibration factors derived from each standard for each filter are then averaged. The pipeline inserts this value and its associated uncertainty into the headers of the Level 2 data files for the flux standards, and uses the value to produce calibrated flux standards. The final step involves examining the calibration values and ensuring that the values are consistent. Outlier values may come from bad observations of a standard star; these values are removed to produce a robust average of the calibration factor across the flight series. The resulting average values are then used to calibrate the observations of the science targets.
Using the telluric-corrected photometry of the standard, \(N_e^{std,corr}\) (in ct/s), and the predicted mean fluxes of the standards in each filter, \(\langle F_{\nu}^{std} \rangle\) (in Jy), the flux of a target object is given by
where \(N_e^{obj,corr}\) is the telluric-corrected count rate in ct/s detected from the source, \(C\) is the calibration factor (ct/s/Jy), and \(F_{\nu}^{nom,obj}(\lambda_{ref})\) is the flux in Jy of a nominal, flat spectrum source (for which \(F_{\nu} \sim \nu^{-1}\)) at a reference wavelength \(\lambda_{ref}\).
The values of \(C\), \(\sigma_C\), and \(\lambda_{ref}\) are written into the headers of the calibrated (PROCSTAT=LEVEL_3 ) data as the keywords CALFCTR, ERRCALF, and LAMREF, respectively. The reference wavelength \(\lambda_{ref}\) for these observations was taken to be the mean wavelengths of the filters, \(\langle \lambda \rangle\).
Note that \(\sigma_C\), as stored in the ERRCALF value, is derived from the standard deviation of the calibration factors across multiple flights. These values are typically on the order of about 6%. There is an additional systematic uncertainty on the stellar models, which is on the order of 3-6%.
The atmospheric transmission in each filter has been computed using the ATRAN code (Lord 1992) for a range of observatory altitudes (corresponding to a range of overhead precipitable water vapor values) and telescope elevations. The ratio of the transmission at each altitude and zenith angle relative to that at the reference altitude (41000 feet) and zenith angle (45 degrees) has been calculated for each filter and fit with a low order polynomial. The ratio appropriate for the altitude and zenith angle of each observation is calculated and applied to each image.
Color corrections¶
An observer often wishes to determine the true flux of an object at the reference wavelength, \(F_{\nu}^{obj}(\lambda_{ref})\), rather than the flux of an equivalent nominal, flat spectrum source. To do this, we define a color correction K such that
where \(F_{\nu}^{nom,obj}(\lambda_{ref})\) is the flux density obtained by measurement on a data product. Divide the measured values by K to obtain the “true” flux density. In terms of the wavelengths defined above,
For most filters and spectral shapes, the color corrections are small (<10%). Tables listing K values and filter wavelengths are available from the SOFIA website.
Spectrophotometric Flux Calibration¶
The common approach to characterizing atmospheric transmission for ground-based infrared spectroscopy is to obtain, for every science target, similar observations of a spectroscopic standard source with as close a match as possible in both airmass and time. Such an approach is not practical for airborne observations, as it imposes too heavy a burden on flight planning and lowers the efficiency of science observations. Therefore, we employ a calibration plan that incorporates a few observations of a calibration star per flight and a model of the atmospheric absorption for the approximate altitude and airmass (and precipitable water vapor, if known) at which the science objects were observed.
Instrumental response curves are generated from the extracted spectra of calibrator targets, typically A0V stars with stellar models constructed from a model of Vega. The extracted spectra are corrected for telluric absorption using the ATRAN models corresponding to the altitude and zenith angle of the calibrator observations, smoothed to the nominal resolution for the grism/slit combination, and sampled at the observed spectral binning. The telluric-corrected spectra are then divided by the appropriate models to generate response curves (with units of ct/s/Jy at each wavelength) for each grism passband. The response curves derived from the various calibrators for each instrumental combination are then combined and smoothed to generate a set of master instrumental response curves. The statistical uncertainties on these response curves are on the order of 5-10%.
Flux calibration of FLITECAM grism data for a science target is currently carried out in a two-step process:
For any given observation of a science target, the closest telluric model (in terms of altitude and airmass of the target observations) is selected and then smoothed to the observed resolution and sampled at the observed spectral binning. The observed spectrum is then divided by the smoothed and re-sampled telluric model.
The telluric-corrected spectrum is then divided by a response function corresponding to the observed instrument mode to convert DN/s to Jy at each pixel.
In order to account for any wavelength shifts between the models and the observations, an optimal shift is estimated by minimizing the residuals of the corrected spectrum, with respect to small relative wavelength shifts between the observed data and the telluric spectrum. This wavelength shift is applied to the data before dividing by the telluric model and response function.
Based on our experience with FORCAST calibration, and with using A0V stars to calibrate near-infrared data, we expect the overall error in the flux calibration to be about 10-20%. However, the uncertainty on the slope of the calibrated spectrum should be substantially less than that, on the order of a few percent (see e.g., Cushing et al. 2005; Rayner et al. 2009). The Level 3 data product for any grism includes the calibrated spectrum and an error spectrum that incorporates these RMS values. The adopted telluric absorption model and the instrumental response functions are also provided in the output product.
As for any slit spectrograph, highly accurate absolute flux levels from FLITECAM grism observations (for absolute spectrophotometry, for example) require additional photometric observations to correct the calibrated spectra for slit losses that can be variable (due to varying image quality) between the spectroscopic observations of the science target and the calibration standard.
Data products¶
Filenames¶
FLITECAM output files from Redux are named according to the convention:
FILENAME = F[flight]_FC_IMA|GRI_AOR-ID_SPECTEL1_Type_FN1[-FN2].fits
where flight is the SOFIA flight number, FC is the instrument identifier, IMA or GRI specifies that it is an imaging or grism file, AOR-ID is the AOR identifier for the observation, SPECTEL1 is the keywords specifying the filter or grism used, Type is three letters identifying the product type (listed in Table 9 and Table 10 below), and FN1 is the file number corresponding to the input file. FN1-FN2 is used if there are multiple input files for a single output file, where FN1 is the file number of the first input file and FN2 is the file number of the last input file.
Pipeline Products¶
The following tables list all intermediate products generated by the pipeline for imaging and grism modes, in the order in which they are produced. [6] The product type is stored in the FITS headers under the keyword PRODTYPE. By default, for imaging, the flat, telluric_corrected, coadded, and calibrated products are saved. For spectroscopy, the spectral_image, rectified_image, spectra, spectra_1d, calibrated_spectrum, coadded_spectrum, and combined_spectrum products are saved.
The final grism mode output product from the Combine Spectra or Combine Response steps are dependent on the input data: for OBSTYPE=STANDARD_TELLURIC, the instrument_response is produced instead of a coadded_spectrum and combined_spectrum.
For most observation modes, the pipeline additionally produces an image in PNG format, intended to provide a quick-look preview of the data contained in the final product. These auxiliary products may be distributed to observers separately from the FITS file products.
Earlier versions of this pipeline (before v2.0.0) produced different sets of default products. Refer to earlier revisions of this manual for complete information.
Step
|
Data type
|
PRODTYPE
|
PROCSTAT
|
Code
|
Saved
|
Extensions
|
|---|---|---|---|---|---|---|
Correct Nonlinearity
|
2D image
|
linearized
|
LEVEL_2
|
LNZ
|
N
|
FLUX, ERROR, BADMASK
|
Clip Image
|
2D image
|
clipped
|
LEVEL_2
|
CLP
|
N
|
FLUX, ERROR,
BADMASK, EXPOSURE
|
Make Flat
|
2D image
|
flat
|
LEVEL_2
|
FLT
|
Y
|
FLUX, ERROR,
BADMASK, EXPOSURE
FLAT, FLAT_ERROR,
FLAT_BADMASK
|
Correct Gain
|
2D image
|
gain_
corrected
|
LEVEL_2
|
GCR
|
N
|
FLUX, ERROR,
BADMASK, EXPOSURE
|
Subtract Sky
|
2D image
|
background_
subtracted
|
LEVEL_2
|
BGS
|
N
|
FLUX, ERROR,
BADMASK, EXPOSURE
|
Register
|
2D image
|
registered
|
LEVEL_2
|
REG
|
N
|
FLUX, ERROR,
BADMASK, EXPOSURE
|
Telluric Correct
|
2D image
|
telluric_
corrected
|
LEVEL_2
|
TEL
|
Y
|
FLUX, ERROR,
BADMASK, EXPOSURE
|
Coadd
|
2D image
|
coadded
|
LEVEL_2
|
COA
|
Y
|
FLUX, ERROR, EXPOSURE
|
Flux Calibrate
|
2D image
|
calibrated
|
LEVEL_3
|
CAL
|
Y
|
FLUX, ERROR, EXPOSURE
|
Mosaic
|
2D image
|
mosaic
|
LEVEL_4
|
MOS
|
Y
|
FLUX, ERROR, EXPOSURE
|
Step
|
Data type
|
PRODTYPE
|
PROCSTAT
|
Code
|
Saved
|
Extensions
|
|---|---|---|---|---|---|---|
Correct Nonlinearity
|
2D spectral
image
|
linearized
|
LEVEL_2
|
LNZ
|
N
|
FLUX, ERROR, BADMASK
|
Make Spectral Image
|
2D spectral
image
|
spectral_
image
|
LEVEL_2
|
IMG
|
Y
|
FLUX, ERROR
|
Stack Dithers
|
2D spectral
image
|
dithers_
stacked
|
LEVEL_2
|
SKD
|
N
|
FLUX, ERROR
|
Make Profiles
|
2D spectral
image
|
rectified_
image
|
LEVEL_2
|
RIM
|
Y
|
FLUX, ERROR, BADMASK,
WAVEPOS, SLITPOS,
SPATIAL_MAP,
SPATIAL_PROFILE
|
Locate Apertures
|
2D spectral
image
|
apertures_
located
|
LEVEL_2
|
LOC
|
N
|
FLUX, ERROR, BADMASK,
WAVEPOS, SLITPOS,
SPATIAL_MAP,
SPATIAL_PROFILE
|
Trace Continuum
|
2D spectral
image
|
continuum_
traced
|
LEVEL_2
|
TRC
|
N
|
FLUX, ERROR, BADMASK,
WAVEPOS, SLITPOS,
SPATIAL_MAP,
SPATIAL_PROFILE,
APERTURE_TRACE
|
Set Apertures
|
2D spectral
image
|
apertures_set
|
LEVEL_2
|
APS
|
N
|
FLUX, ERROR, BADMASK,
WAVEPOS, SLITPOS,
SPATIAL_MAP,
SPATIAL_PROFILE,
APERTURE_TRACE,
APERTURE_MASK
|
Subtract
Background
|
2D spectral
image
|
background_
subtracted
|
LEVEL_2
|
BGS
|
N
|
FLUX, ERROR, BADMASK,
WAVEPOS, SLITPOS,
SPATIAL_MAP,
SPATIAL_PROFILE,
APERTURE_TRACE,
APERTURE_MASK
|
Extract Spectra
|
2D spectral
image;
1D spectrum
|
spectra
|
LEVEL_2
|
SPM
|
Y
|
FLUX, ERROR, BADMASK,
WAVEPOS, SLITPOS,
SPATIAL_MAP,
SPATIAL_PROFILE,
APERTURE_TRACE,
APERTURE_MASK,
SPECTRAL_FLUX,
SPECTRAL_ERROR,
TRANSMISSION
|
Extract Spectra
|
1D spectrum
|
spectra_1d
|
LEVEL_3
|
SPC
|
Y
|
FLUX
|
Calibrate Flux
|
2D spectral
image;
1D spectrum
|
calibrated_
spectrum
|
LEVEL_3
|
CRM
|
Y
|
FLUX, ERROR, BADMASK,
WAVEPOS, SLITPOS,
SPATIAL_MAP,
SPATIAL_PROFILE,
APERTURE_TRACE,
APERTURE_MASK,
SPECTRAL_FLUX,
SPECTRAL_ERROR
TRANSMISSION,
RESPONSE,
RESPONSE_ERROR
|
Combine Spectra
|
2D spectral
image;
1D spectrum
|
coadded_
spectrum
|
LEVEL_3
|
COA
|
Y
|
FLUX, ERROR,
EXPOSURE, WAVEPOS,
SPECTRAL_FLUX,
SPECTRAL_ERROR
TRANSMISSION,
RESPONSE
|
Combine Spectra
|
1D spectrum
|
combined_
spectrum
|
LEVEL_3
|
CMB
|
Y
|
FLUX
|
Make Response
|
1D response
spectrum
|
response_
spectrum
|
LEVEL_3
|
RSP
|
Y
|
FLUX
|
Combine Response
|
1D response
spectrum
|
instrument_
response
|
LEVEL_4
|
IRS
|
Y
|
FLUX
|
Data Format¶
All files produced by the pipeline are multi-extension FITS files (except for the combined_spectrum, response_spectrum, and instrument_response products: see below). [7] The flux image is stored in the primary header-data unit (HDU); its associated error image is stored in extension 1, with EXTNAME=ERROR.
Imaging products may additionally contain an extension with EXTNAME=EXPOSURE, which contains the nominal exposure time at each pixel, in seconds. This extension has the same meaning for the spectroscopic coadded_spectrum product.
In spectroscopic products, the SLITPOS and WAVEPOS extensions give the spatial (rows) and spectral (columns) coordinates, respectively, for rectified images. These coordinates may also be derived from the WCS in the primary header. WAVEPOS also indicates the wavelength coordinates for 1D extracted spectra.
Intermediate spectral products may contain SPATIAL_MAP and SPATIAL_PROFILE extensions. These contain the spatial map and median spatial profile, described in the Rectify spectral image section, above. They may also contain APERTURE_TRACE and APERTURE_MASK extensions. These contain the spectral aperture definitions, as described in the Identify apertures section.
Final spectral products contain SPECTRAL_FLUX and SPECTRAL_ERROR extensions: these are the extracted 1D spectrum and associated uncertainty. They also contain TRANSMISSION and RESPONSE extensions, containing the atmospheric transmission and instrumental response spectra used to calibrate the spectrum (see the Calibrate flux and correct for atmospheric transmission section).
The combined_spectrum, response_spectrum, and instrument_response are one-dimensional spectra, stored in Spextool format, as rows of data in the primary extension.
For the combined_spectrum, the first row is the wavelength (um), the second is the flux (Jy), the third is the error (Jy), the fourth is the estimated fractional atmospheric transmission spectrum, and the fifth is the instrumental response curve used in flux calibration (ct/s/Jy). These rows correspond directly to the WAVEPOS, SPECTRAL_FLUX, SPECTRAL_ERROR, TRANSMISSION, and RESPONSE extensions in the coadded_spectrum product.
For the response_spectrum, generated from telluric standard observations, the first row is the wavelength (um), the second is the response spectrum (ct/s/Jy), the third is the error on the response (ct/s/Jy), the fourth is the atmospheric transmission spectrum (unitless), and the fifth is the standard model used to derive the response (Jy). The instrument_reponse spectrum, generated from combined response_spectrum files, similarly has wavelength (um), response (ct/s/Jy), error (ct/s/Jy), and transmission (unitless) rows.
The final uncertainties in calibrated images and spectra contain only the estimated statistical uncertainties due to the noise in the image or the extracted spectrum. The systematic uncertainties due to the calibration process are recorded in header keywords. For imaging data, the error on the calibration factor is recorded in the keyword ERRCALF. For grism data, the estimated overall fractional error on the flux is recorded in the keyword CALERR. [8]
In earlier versions of this pipeline (prior to 2.0.0), all image products were 3D arrays of data, where the first plane was the image and the second plane was the error associated with each pixel in the image. An optional third plane was the bad pixel mask, indicating the location of bad pixels. All spectral products were in the Spextool format described above for the combined_spectrum product.
Earlier versions of this pipeline (prior to 2.0.0) may have stored the systematic calibration error in the error spectrum or variance image, added in quadrature with the statistical error. Check PIPEVERS and compare the error estimates for the calibrated products to earlier products to ensure correct interpretation of the error estimates.
Grouping LEVEL_1 data for processing¶
For both imaging and grism mode for FLITECAM, there are two possible kinds of input data: sky frames and sources. Sky frames have OBSTYPE = SKY, while source frames may have OBSTYPE = OBJECT, STANDARD_FLUX, or STANDARD_TELLURIC. The sky frames and source frames should all share the same instrument configuration and filters. Optionally, it may also be useful to separate out data files taken from different missions, observation plans, or AOR-IDs.
These grouping requirements translate into a set of FITS keywords that must match in order for a set of data to be grouped together. These relationships are summarized in the tables below.
Note that data grouping must be carried out before the pipeline is run. The pipeline expects all inputs to be reduced together as a single data set.
Keyword |
Data Type |
Match Criterion |
|---|---|---|
OBSTYPE |
STR |
Exact (unless SKY) |
OBJECT |
STR |
Exact |
INSTCFG |
STR |
Exact |
INSTMODE |
STR |
Exact |
SPECTEL1 |
STR |
Exact |
MISSN-ID (optional) |
STR |
Exact |
PLANID (optional) |
STR |
Exact |
AOR_ID (optional) |
STR |
Exact |
Keyword |
Data Type |
Match Criterion |
|---|---|---|
OBSTYPE |
STR |
Exact (unless SKY) |
OBJECT |
STR |
Exact |
INSTCFG |
STR |
Exact |
INSTMODE |
STR |
Exact |
SPECTEL1 |
STR |
Exact |
SPECTEL2 |
STR |
Exact |
MISSN-ID (optional) |
STR |
Exact |
PLANID (optional) |
STR |
Exact |
AOR_ID (optional) |
STR |
Exact |
Configuration and execution¶
Installation¶
The FLITECAM pipeline is written entirely in Python. The pipeline is platform independent and has been tested on Linux, Mac OS X, and Windows operating systems. Running the pipeline requires a minimum of 16GB RAM, or equivalent-sized swap file.
The pipeline is comprised of six modules within the sofia_redux package:
sofia_redux.instruments.flitecam, sofia_redux.instruments.forcast,
sofia_redux.pipeline, sofia_redux.calibration, sofia_redux.spectroscopy,
sofia_redux.toolkit, and sofia_redux.visualization.
The flitecam module provides the data processing
algorithms specific to FLITECAM, with supporting libraries from the
forcast, toolkit, calibration, spectroscopy, and visualization
modules. The pipeline module provides interactive and batch interfaces
to the pipeline algorithms.
External Requirements¶
To run the pipeline for any mode, Python 3.8 or
higher is required, as well as the following packages: numpy, scipy,
matplotlib, pandas, astropy, configobj, numba, bottleneck, joblib,
and photutils.
Some display functions for the graphical user interface (GUI)
additionally require the PyQt5, pyds9, and regions packages.
All required external packages are available to install via the
pip or conda package managers. See the Anaconda environment file
(environment.yml), or the pip requirements file (requirements.txt)
distributed with sofia_redux for up-to-date version requirements.
Running the pipeline interactively also requires an installation of SAO DS9 for FITS image display. See http://ds9.si.edu/ for download and installation instructions. The ds9 executable must be available in the PATH environment variable for the pyds9 interface to be able to find and control it. Please note that pyds9 is not available on the Windows platform.
Source Code Installation¶
The source code for the FLITECAM pipeline maintained by the SOFIA Data Processing Systems (DPS) team can be obtained directly from the DPS, or from the external GitHub repository. This repository contains all needed configuration files, auxiliary files, and Python code to run the pipeline on FLITECAM data in any observation mode.
After obtaining the source code, install the package with the command:
python setup.py install
from the top-level directory.
Alternately, a development installation may be performed from inside the directory with the command:
pip install -e .
After installation, the top-level pipeline interface commands should be available in the PATH. Typing:
redux
from the command line should launch the GUI interface, and:
redux_pipe -h
should display a brief help message for the command line interface.
Configuration¶
For FLITECAM algorithms, default parameter values are defined by the Redux object that interfaces to them. These values may be overridden manually for each step, while running in interactive mode. They may also be overridden by an input parameter file, in INI format, in either interactive or automatic mode. See Appendix A for an example of an input parameter file, which contains the current defaults for all parameters.
Input data¶
Redux takes as input raw FLITECAM FITS data files containing 1024x1024 pixel image arrays. The FITS headers contain data acquisition and observation parameters and, combined with the pipeline configuration files, comprise the information necessary to complete all steps of the data reduction process. Some critical keywords are required to be present in the raw data in order to perform a successful grouping, reduction, and ingestion into the SOFIA archive. See Appendix B for a description of these keywords.
It is assumed that the input data have been successfully grouped before beginning reduction: Redux considers all input files in a reduction to be part of a single homogeneous reduction group, to be reduced together with the same parameters. As such, when the pipeline reads a raw FLITECAM data file, it uses the first input file to identify the observing mode used. Given this information, it identifies a set of auxiliary and calibration data files to be used in the reduction (Table 13). The default files to be used are defined in a lookup table that reads the DATE-OBS keyword from the raw file, and then chooses the appropriate calibrations for that date.
Auxiliary data file |
Data type |
Comments |
|---|---|---|
Keyword definition file (e.g. header_req_ima.cfg) |
INI |
Contains the definition of required keywords, with allowed value ranges |
Linearity coefficients file (e.g. lc_coeffs_20140325.fits) |
FITS |
FITS file containing linearity correction coefficients for all pixels in the raw data array |
Reference flux calibration table (e.g. refcalfac_20171007.txt) |
ASCII |
Flux calibration factors (imaging only) |
Spectral order definition file (e.g. flat_kla_multinight.fits) |
FITS |
Image file containing flat data and an edge mask for a spectral order (grism only) |
Wavelength calibration map (e.g. kla_map_arcs.fits) |
FITS |
Two-frame image associating a wavelength value and a spatial distance across the slit with each pixel (grism only) |
Atmospheric transmission curve (e.g. atran_41K_45deg_1-6mum.fits) |
FITS |
A FITS image with wavelength and transmission values for a particular altitude and zenith angle (grism only) |
Instrumental response curve (e.g. FC_GRI_A2KL_SS20_RSP.fits) |
FITS |
FITS image containing the response at each wavelength for a particular grism/slit mode (grism only) |
Automatic Mode Execution¶
The DPS pipeline infrastructure runs a pipeline on previously-defined reduction groups as a fully-automatic black box. To do so, it creates an input manifest (infiles.txt) that contains relative paths to the input files (one per line). The command-line interface to the pipeline is run as:
redux_pipe infiles.txt
The command-line interface will read in the specified input files, use their headers to determine the observation mode, and accordingly the steps to run and any intermediate files to save. Output files are written to the current directory, from which the pipeline was called. After reduction is complete, the script will generate an output manifest (outfiles.txt) containing the relative paths to all output FITS files generated by the pipeline.
Optionally, in place of a manifest file, file paths to input files may be directly specified on the command line. Input files may be raw FITS files, or may be intermediate products previously produced by the pipeline. For example, this command will complete the reduction for a set of FITS files in the current directory, previously reduced through the calibration step of the pipeline:
redux_pipe *CAL*.fits
To customize batch reductions from the command line, the redux_pipe interface accepts a configuration file on the command line. This file may contain any subset of the full configuration file, specifying any non-default parameters for pipeline steps. An output directory for pipeline products and the terminal log level may also be set on the command line.
The full set of optional command-line parameters accepted by the redux_pipe interface are:
-h, --help show this help message and exit
-c CONFIG, --configuration CONFIG
Path to Redux configuration file.
-o OUTDIR, --out OUTDIR
Path to output directory.
-l LOGLEVEL, --loglevel LOGLEVEL
Log level.
Manual Mode Execution¶
In manual mode, the pipeline may be run interactively, via a graphical user interface (GUI) provided by the Redux package. The GUI is launched by the command:
redux
entered at the terminal prompt (Fig. 53). The GUI allows output directory specification, but it may write initial or temporary files to the current directory, so it is recommended to start the interface from a location to which the user has write privileges.
From the command line, the redux interface accepts an optional config file (-c) or log level specification (-l), in the same way the redux_pipe command does. Any pipeline parameters provided to the interface in a configuration file will be used to set default values; they will still be editable from the GUI.
Fig. 53 Redux GUI startup.¶
Basic Workflow¶
To start an interactive reduction, select a set of input files, using the File menu (File->Open New Reduction). This will bring up a file dialog window (see Fig. 54). All files selected will be reduced together as a single reduction set.
Redux will decide the appropriate reduction steps from the input files, and load them into the GUI, as in Fig. 55.
Fig. 54 Open new reduction.¶
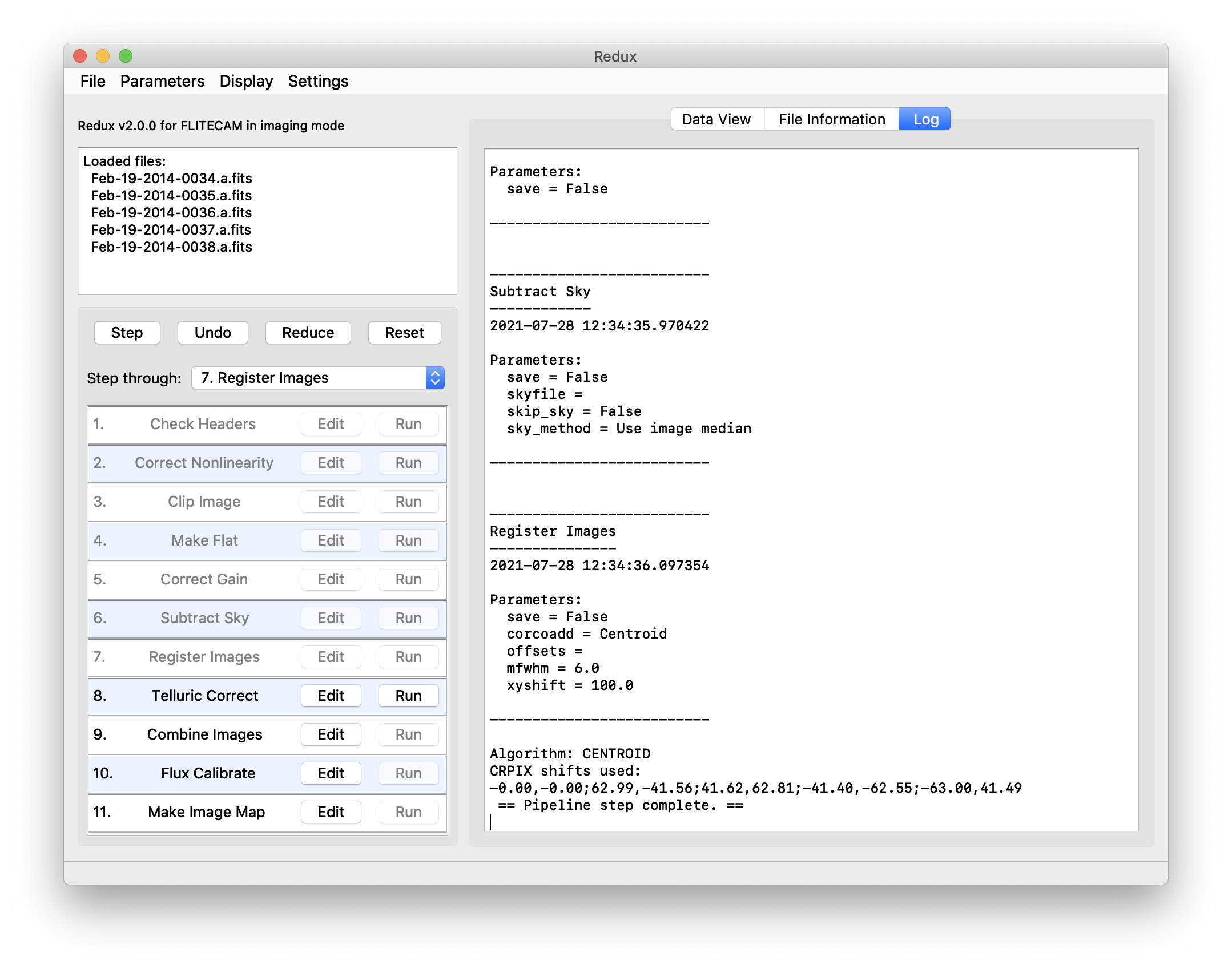
Fig. 55 Sample reduction steps. Log output from the pipeline is displayed in the Log tab.¶
Each reduction step has a number of parameters that can be edited before running the step. To examine or edit these parameters, click the Edit button next to the step name to bring up the parameter editor for that step (Fig. 56). Within the parameter editor, all values may be edited. Click OK to save the edited values and close the window. Click Reset to restore any edited values to their last saved values. Click Restore Defaults to reset all values to their stored defaults. Click Cancel to discard all changes to the parameters and close the editor window.
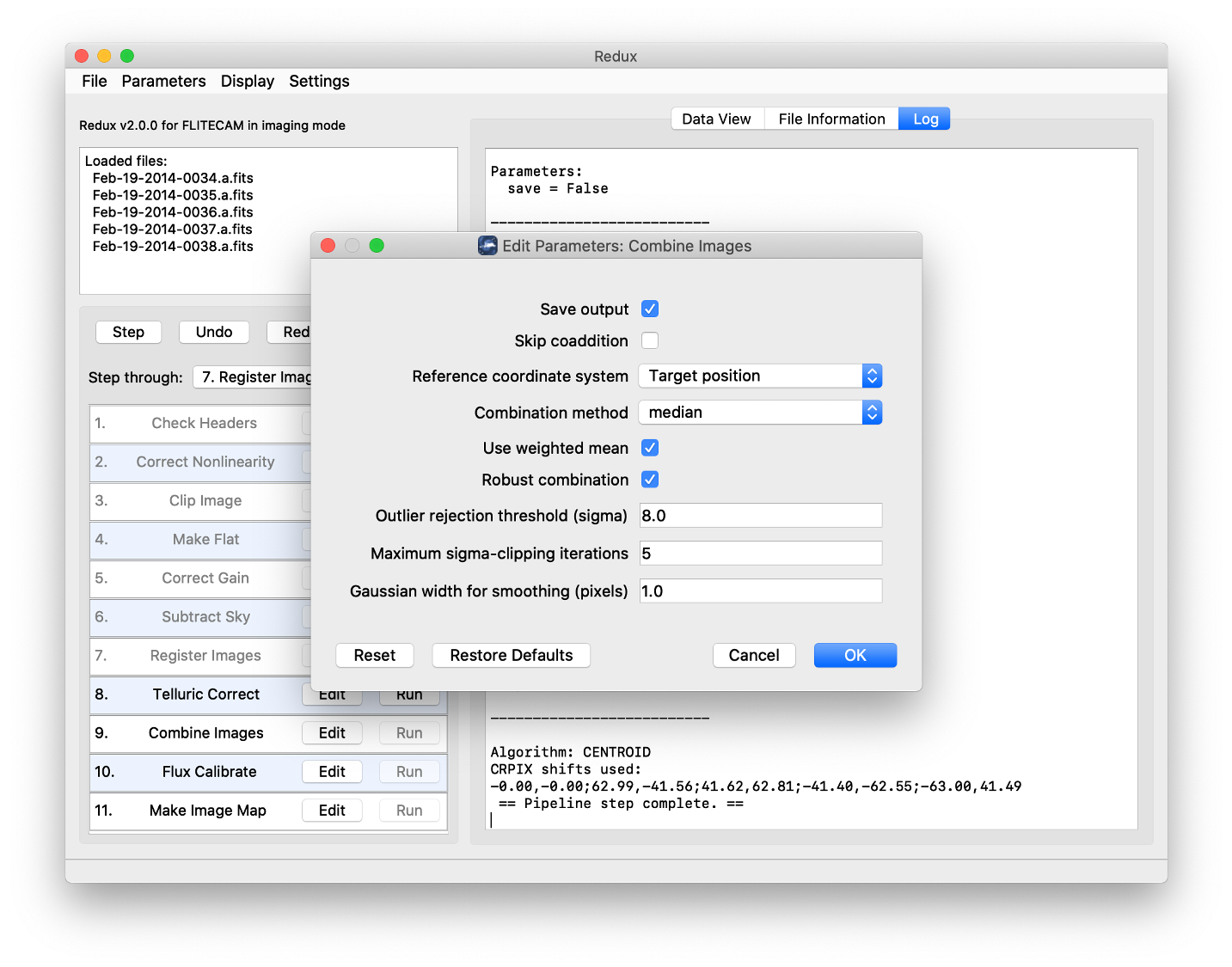
Fig. 56 Sample parameter editor for a pipeline step.¶
The current set of parameters can be displayed, saved to a file, or reset all at once using the Parameters menu. A previously saved set of parameters can also be restored for use with the current reduction (Parameters -> Load Parameters).
After all parameters for a step have been examined and set to the user’s satisfaction, a processing step can be run on all loaded files either by clicking Step, or the Run button next to the step name. Each processing step must be run in order, but if a processing step is selected in the Step through: widget, then clicking Step will treat all steps up through the selected step as a single step and run them all at once. When a step has been completed, its buttons will be grayed out and inaccessible. It is possible to undo one previous step by clicking Undo. All remaining steps can be run at once by clicking Reduce. After each step, the results of the processing may be displayed in a data viewer. After running a pipeline step or reduction, click Reset to restore the reduction to the initial state, without resetting parameter values.
Files can be added to the reduction set (File -> Add Files) or removed from the reduction set (File -> Remove Files), but either action will reset the reduction for all loaded files. Select the File Information tab to display a table of information about the currently loaded files (Fig. 57).
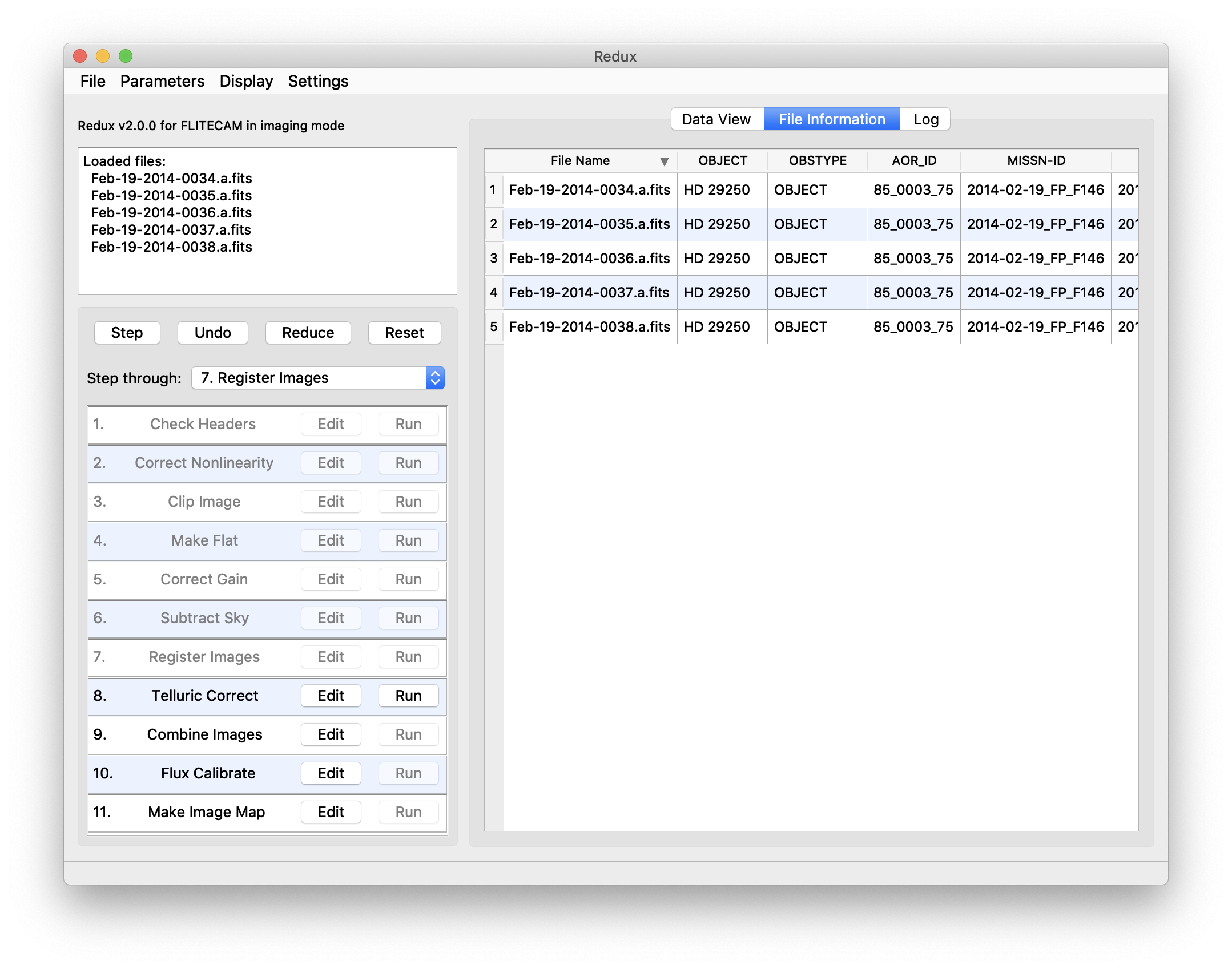
Fig. 57 File information table.¶
Display Features¶
The Redux GUI displays images for quality analysis and display (QAD) in the DS9 FITS viewer. DS9 is a standalone image display tool with an extensive feature set. See the SAO DS9 site (http://ds9.si.edu/) for more usage information.
After each pipeline step completes, Redux may load the produced images into DS9. Some display options may be customized directly in DS9; some commonly used options are accessible from the Redux interface, in the Data View tab (Fig. 58).
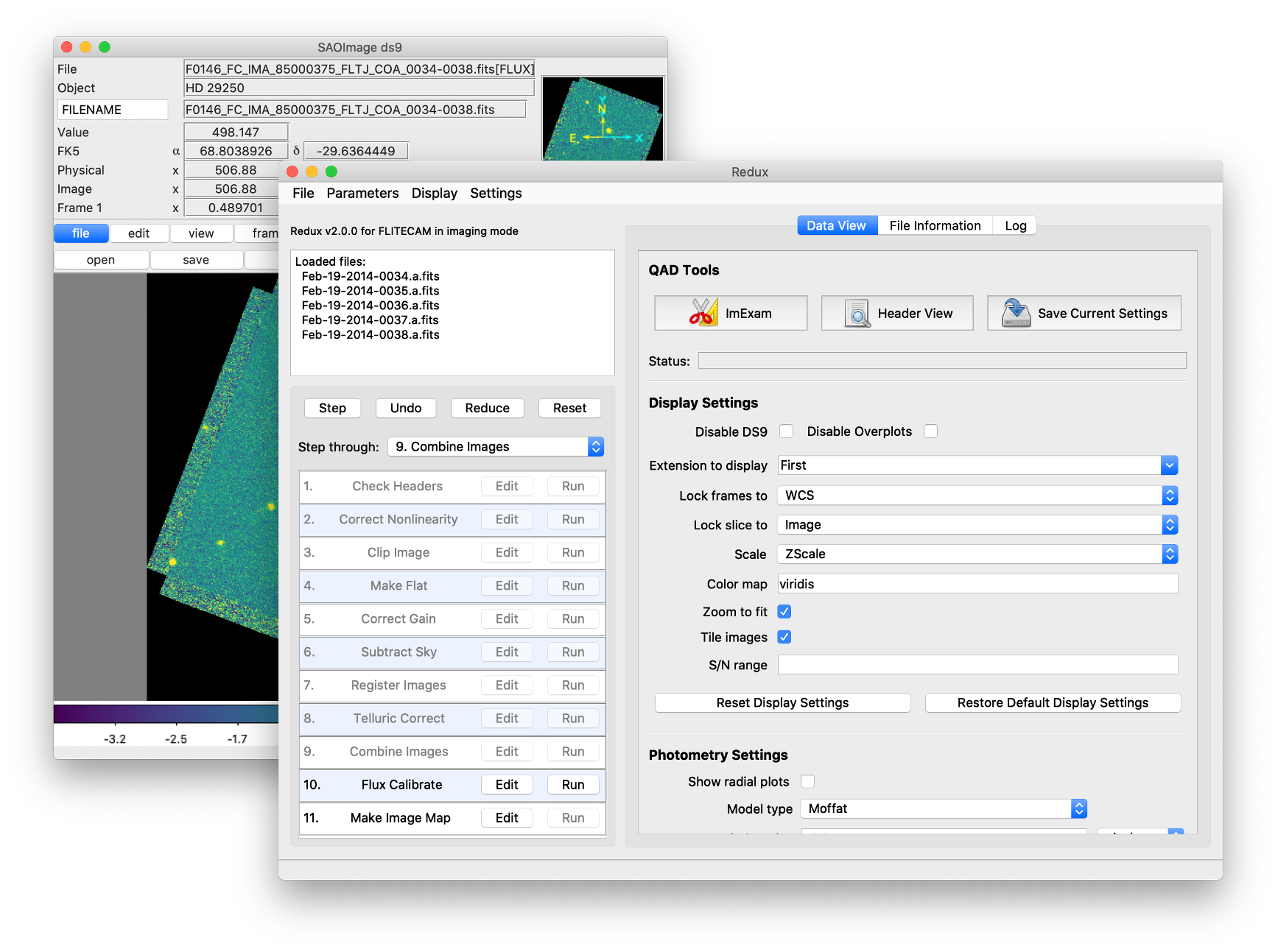
Fig. 58 Data viewer settings and tools.¶
From the Redux interface, the Display Settings can be used to:
Set the FITS extension to display (First, or edit to enter a specific extension), or specify that all extensions should be displayed in a cube or in separate frames.
Lock individual frames together, in image or WCS coordinates.
Lock cube slices for separate frames together, in image or WCS coordinates.
Set the image scaling scheme.
Set a default color map.
Zoom to fit image after loading.
Tile image frames, rather than displaying a single frame at a time.
Changing any of these options in the Data View tab will cause the currently displayed data to be reloaded, with the new options. Clicking Reset Display Settings will revert any edited options to the last saved values. Clicking Restore Default Display Settings will revert all options to their default values.
In the QAD Tools section of the Data View tab, there are several additional tools available.
Clicking the ImExam button (scissors icon) launches an event loop in DS9. After launching it, bring the DS9 window forward, then use the keyboard to perform interactive analysis tasks:
Type ‘a’ over a source in the image to perform photometry at the cursor location.
Type ‘p’ to plot a pixel-to-pixel comparison of all frames at the cursor location.
Type ‘s’ to compute statistics and plot a histogram of the data at the cursor location.
Type ‘c’ to clear any previous photometry results or active plots.
Type ‘h’ to print a help message.
Type ‘q’ to quit the ImExam loop.
The photometry settings (the image window considered, the model fit,
the aperture sizes, etc.) may be customized in the Photometry Settings.
Plot settings (analysis window size, shared plot axes, etc.) may be
customized in the Plot Settings.
After modifying these settings, they will take effect only for new
apertures or plots (use ‘c’ to clear old ones first). As for the display
settings, the reset button will revert to the last saved values
and the restore button will revert to default values.
For the pixel-to-pixel and histogram plots, if the cursor is contained within
a previously defined DS9 region (and the regions package is installed),
the plot will consider only pixels within the region. Otherwise, a window
around the cursor is used to generate the plot data. Setting the window
to a blank value in the plot settings will use the entire image.
Clicking the Header button (magnifying glass icon) from the QAD Tools section opens a new window that displays headers from currently loaded FITS files in text form (Fig. 59). The extensions displayed depends on the extension setting selected (in Extension to Display). If a particular extension is selected, only that header will be displayed. If all extensions are selected (either for cube or multi-frame display), all extension headers will be displayed. The buttons at the bottom of the window may be used to find or filter the header text, or generate a table of header keywords. For filter or table display, a comma-separated list of keys may be entered in the text box.
Clicking the Save Current Settings button (disk icon) from the QAD Tools section saves all current display and photometry settings for the current user. This allows the user’s settings to persist across new Redux reductions, and to be loaded when Redux next starts up.
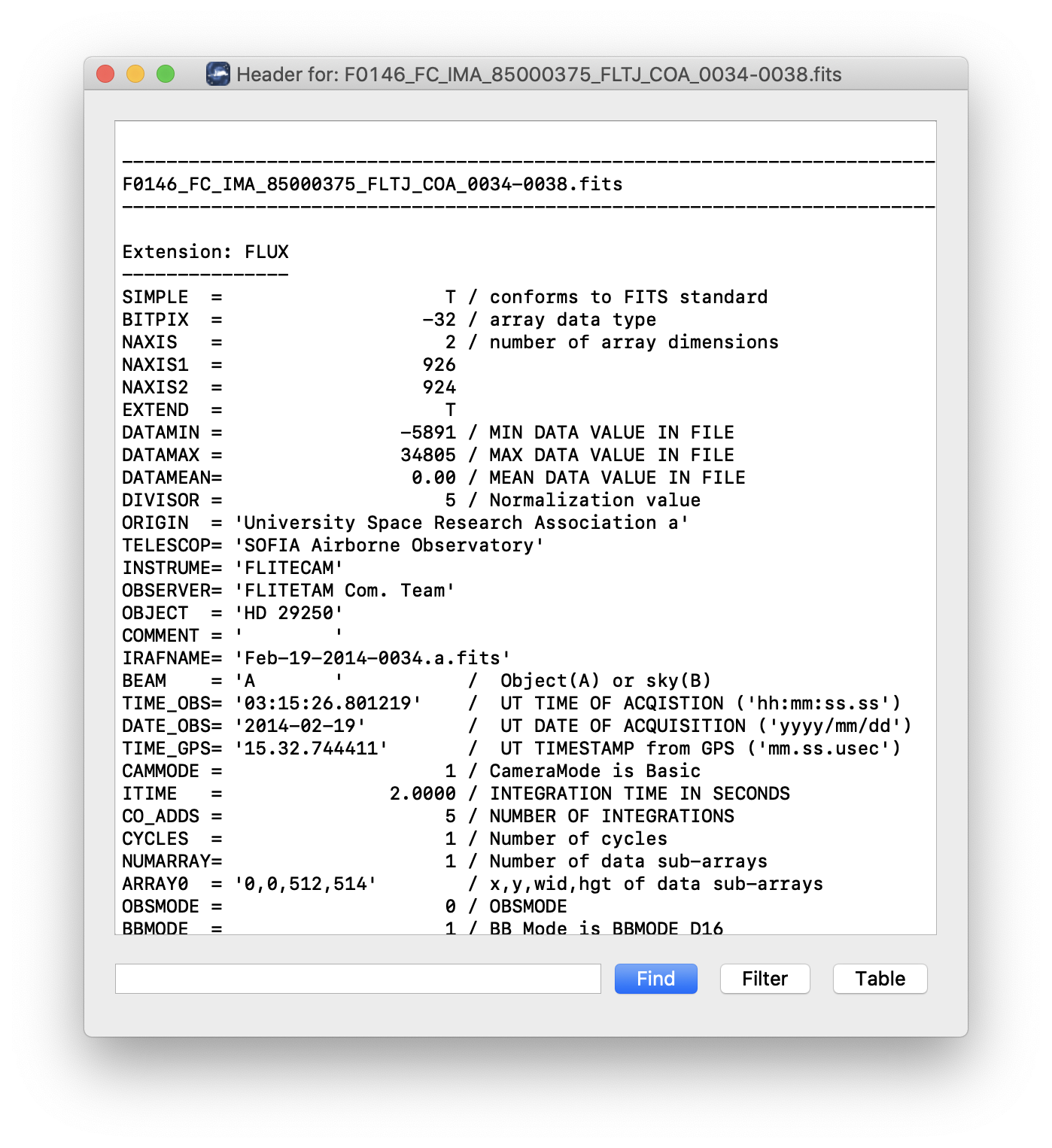
Fig. 59 QAD FITS header viewer.¶
FLITECAM Reduction¶
Imaging Reduction¶
FLITECAM imaging reduction with Redux is straightforward. The processing steps follow the flowchart of Fig. 45. At each step, Redux attempts to determine automatically the correct action, given the input data and default parameters, but each step can be customized as needed.
Useful Parameters¶
Some key parameters to note are listed below.
In addition to the specified parameters, the output from each step may be optionally saved by selecting the ‘save’ parameter.
Check Headers
Abort reduction for invalid headers: By default, Redux will halt the reduction if the input header keywords do not meet requirements. Uncheck this box to attempt the reduction anyway.
Correct Nonlinearity:
Linearity correction file: The default linearity correction file on disk is automatically loaded. Set to a valid FITS file path to override the default coefficients file with a new one.
Saturation level: Pixels with raw flux values greater than this value, divided by the DIVISOR, are marked as bad pixels. Set to blank to propagate all pixels, regardless of value.
Clip Image:
Skip clean: If selected, bad pixels will not be identified in the clipped image.
Data to clip to: Enter a pixel range to use as the data section. Values should be entered as xmin, xmax, ymin, ymax, with index values starting at 0. Max values are not included.
Make Flat:
Override flat file: If specified, the provided FITS file will be used in place of generating a flat field from the input data. The provided file must have a FLAT extension. If FLAT_ERROR is also present, the errors will be propagated in the gain correction step.
Skip gain correction: If selected, no flat will be generated, and any file specified as an override will be ignored. Data will not be gain corrected.
Subtract Sky:
Override sky file: If specified, the provided FITS file will be used as a sky image to subtract, in place of using the method parameter. The provided file must have a FLUX extension. If an ERROR extension is also present, the errors will be propagated.
Skip sky subtraction: If selected, no sky values will be subtracted.
Method for deriving sky value: If ‘Use image median’, the background will be determined from the median of each frame. If ‘Use flat normalization value’, the value in the FITS keyword FLATNORM will be subtracted. This is only appropriate if the flat was generated from sky data close in time to the observation (e.g the input data itself).
Register Images
Registration algorithm: The default for all data is to use the WCS as is for registration. Centroiding is may be useful for bright, compact objects; cross-correlation may be useful for bright, diffuse fields. Registration via the ‘Header shifts’ method may be useful for older data, for which the relative WCS is not very accurate. The ‘Use first WCS’ option will treat all images as pre-registered: the data will be coadded directly without shifts.
Override offsets for all images: If registration offsets are known a priori, they may be directly entered here. Specify semi-colon separated offsets, as x,y. For example, for three input images, specify ‘0,0;2,0;0,2’ to leave the first as is, shift the second two pixels to the right in x, and shift the third two pixels up in y.
Expected FWHM for centroiding: Specify the expected FWHM in pixels, for the centroiding algorithm. This may be useful in registering bright compact sources that are not point sources.
Maximum shift for cross-correlation: Specify the maximum allowed shift in pixels for the cross-correlation algorithm. This limit is applied for shifts in both x- and y-direction.
Combine images
Skip coaddition: If selected, each input registered file will be saved as a separate file of type ‘coadded’ rather than combined together into a single output file.
Reference coordinate system: If set to ‘First image’, all images will be referenced to the sky position in the first image file. If set to ‘Target position’, the TGTRA/TGTDEC keywords in the FITS header will be used to apply an additional offset for registering non-sidereal targets. If these keywords are not present, or if their value is constant, the algorithm defaults to the ‘First image’ behavior. ‘Target position’ is on by default; ‘First image’ is recommended only if the TGTRA/TGTDEC keywords are known to have bad values.
Combination method: Median is the default; mean may also be useful for some input data. The resample option may project data more accurately, and allows an additional smoothing option, but takes much longer to complete.
Use weighted mean: If set, the average of the data will be weighted by the variance. Ignored for method=median.
Robust combination: If set, data will be sigma-clipped before combination for mean or median methods.
Outlier rejection threshold (sigma): The sigma-clipping threshold for robust combination methods, in units of sigma (standard deviation).
Gaussian width for smoothing (pixels): If method=resample, a smoothing may be applied to the output averages. Specify the Gaussian width in pixels. Set smaller (but non-zero) for less smoothing; higher for more smoothing.
Flux Calibrate
Re-run photometry for standards: If selected, and observation is a flux standard, photometric fits and aperture measurements on the brightest source will be recalculated, using the input parameters below.
Source position: Enter the approximate position (x,y) of the source to measure. If not specified, the SRCPOSX/SRCPOSY keywords in the FITS header will be used as the first estimate of the source position.
Photometry fit size: Smaller subimages may sometimes be necessary for faint sources and/or variable background.
Initial FWHM: Specify in pixels. This parameter should be modified only if the PSF of the source is significantly larger or smaller than usual.
Profile type: Moffat fits are the default, as they generally give a more reliable FWHM value. However, Gaussian fits may sometimes be more stable, and therefore preferable if the Moffat fit fails.
Make Image Map
Color map: Color map for the output PNG image. Any valid Matplotlib name may be specified.
Flux scale for image: A low and high percentile value , used for scaling the image, e.g. [0,99].
Number of contours: Number of contour levels to be over-plotted on the image. Set to 0 to turn off contours.
Contour color: Color for the contour lines. Any valid Matplotlib color name may be specified.
Filled contours: If set, contours will be filled instead of overlaid.
Overlay grid: If set, a coordinate grid will be overlaid.
Beam marker: If set, a beam marker will be added to the plot.
Watermark text: If set to a non-empty string, the text will be added to the lower-right of the image as a semi-transparent watermark.
Crop NaN border: If set, any remaining NaN or zero-valued border will be cropped out of plot.
Grism Reduction¶
Spectral extraction with Redux is slightly more complicated than image processing. The GUI breaks down the spectral extraction algorithms into six separate reduction steps to give more control over the extraction process. These steps are:
Make Profiles: Generate a smoothed model of the relative distribution of the flux across the slit (the spatial profile). After this step is run, a separate display window showing a plot of the spatial profile appears.
Locate Apertures: Use the spatial profile to identify spectra to extract. By default, Redux attempts to automatically identify sources, but they can also be manually identified by entering a guess position to fit near, or a fixed position, in the parameters. Aperture locations are plotted in the profile window.
Trace Continuum: Identify the location of the spectrum across the array, by either fitting the continuum or fixing the location to the aperture center. The aperture trace is displayed as a region overlay in DS9.
Set Apertures: Identify the data to extract from the spatial profile. This is done automatically by default, but all aperture parameters can be overridden manually in the parameters for this step. Aperture radii and background regions are plotted in the profile window (see Fig. 60).
Subtract Background: Residual background is fit and removed for each column in the 2D image, using background regions specified in the Set Apertures step.
Extract Spectra: Extract one-dimensional spectra from the identified apertures. By default, Redux will perform standard extraction for observations that are marked as extended sources (SRCTYPE=EXTENDED_SOURCE) and will attempt optimal extraction for any other value. The method can be overridden in the parameters for this step.
Extracted spectra are displayed in an interactive plot window, for data analysis and visualization (see Fig. 61).
The spectral display tool has a number of useful features and controls. See Fig. 62 and the table below for a quick summary.
Feature |
Control |
Keyboard shortcut |
|---|---|---|
Load new FITS file |
File Choice -> Add File |
– |
Remove loaded FITS file |
File Choice -> Remove File |
Press delete in the File Choice panel |
Plot selected file |
File Choice -> (double-click) |
– |
Add a new plot window (pane) |
Panes -> Add Pane |
– |
Remove a pane |
Panes -> Remove Pane |
Press delete in the Panes panel, or in the plot window |
Show or hide a plot |
Panes -> Pane # -> File name -> Enabled, or click the Hide all/ Show all button |
– |
Display a different X or Y field (e.g. spectral error, transmission, or response) |
Axis -> X Property or Y Property |
– |
Overplot a different Y axis field (e.g. spectral error, transmission, or response) |
Axis -> Overplot -> Enabled |
– |
Change X or Y units |
Axis -> X Unit or Y Unit |
– |
Change X or Y scale |
Axis -> X Scale or Y Scale -> Linear or Log |
– |
Interactive zoom |
Axis -> Zoom: X, Y, Box, then click in the plot to set the limits; or Reset to reset the limits to default. |
In the plot window, press x, y, or z to start zoom mode in x-direction, y-direction, or box mode, respectively. Click twice on the plot to set the new limits. Press w to reset the plot limits to defaults. |
Fit a spectral feature |
– |
In the plot window, press f to start the fitting mode. Click twice on the plot to set the data limits to fit. |
Change the feature or baseline fit model |
Analysis -> Feature, Background |
– |
Load a spectral line list for overplot display |
Analysis -> Reference Data: Open, Load List Select a one- or two- column text file containing wavelengths in microns and (optionally) labels for the values. Columns may be comma, space, or ‘|’ separated. |
– |
Clear zoom or fit mode |
– |
In the plot window, press c to clear guides and return to default display mode. |
Change the plot color cycle |
Plot -> Color cycle -> Accessible, Spectral or Tableau |
– |
Change the plot type |
Plot -> Plot type -> Step, Line, or Scatter |
– |
Change the plot display options |
Plot -> Show markers, Show errors, Show grid, or Dark mode |
– |
Display the cursor position |
Cursor panel -> Check Cursor Location for a quick display or press Popout for full information |
– |
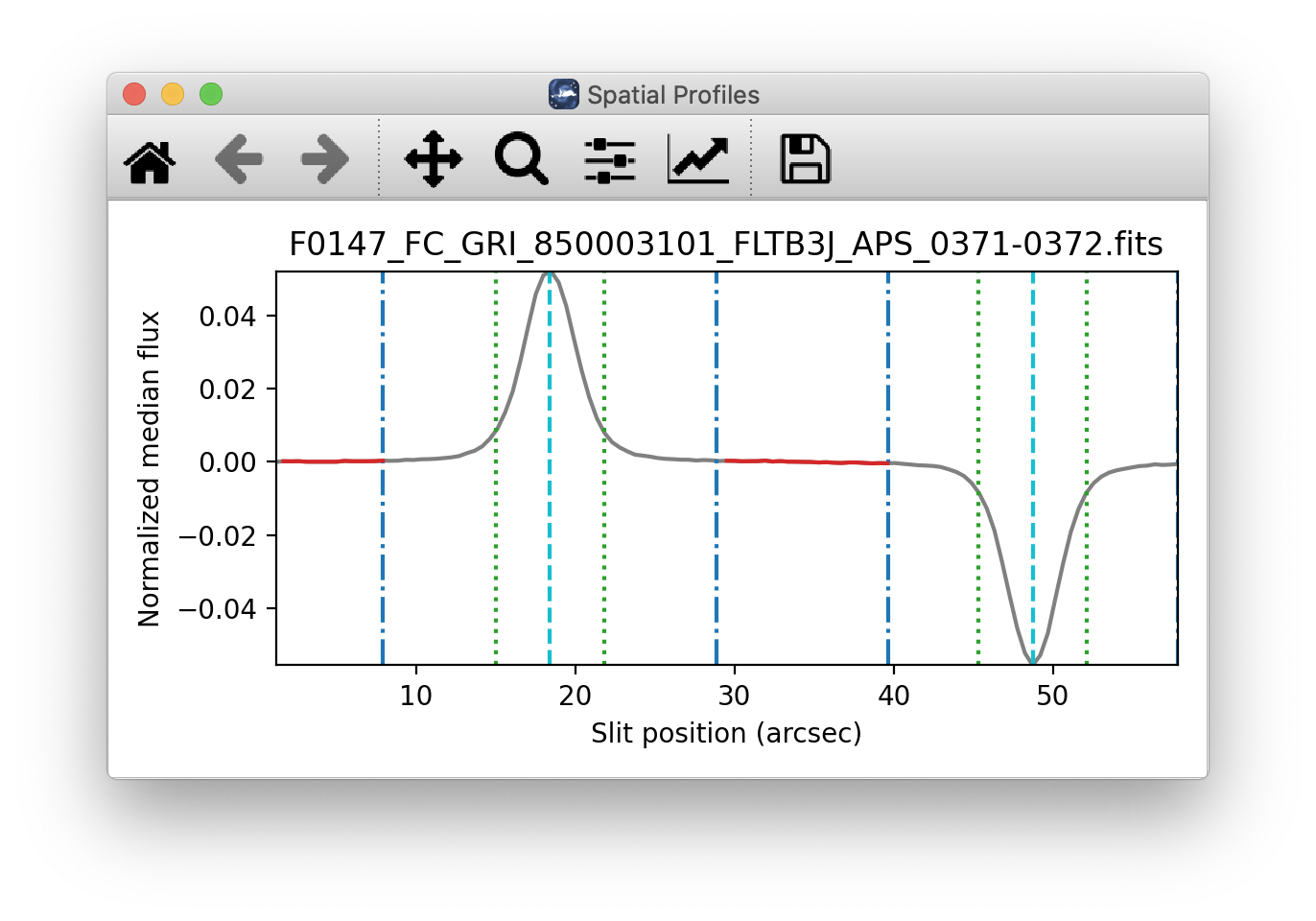
Fig. 60 Aperture location automatically identified and over-plotted on the spatial profile. The cyan line indicates the aperture center. Green lines indicate the integration aperture for optimal extraction, dark blue lines indicate the PSF radius (the point at which the flux goes to zero), and red lines indicate background regions.¶
Fig. 61 Final extracted spectrum, displayed in an interactive plot window.¶
Fig. 62 Control panels for the spectral viewer are located to the left and below the plot window. Click the arrow icons to show or collapse them.¶
Useful Parameters¶
Below are listed some key parameters for the grism processing steps. Note that the Check Headers and Correct Nonlinearity steps are identical to those used for the imaging data: their parameters are listed above. In addition to the specified parameters, the output from each step may be optionally saved by selecting the ‘save’ parameter.
Make Spectral Image:
Subtract pairs: If checked, files will be pair-subtracted in the order they were taken, according to the DATE-OBS keyword in their headers. Mismatched pairs will be dropped from the reduction.
Flat file: If provided, data will be divided by the image in the first extension of the FITS file. Set blank to skip flat correction. The default order mask is automatically loaded for this purpose, but most of the masks provided have flat values set to 1.0, so there is no effective flat correction.
Stack Dithers
Skip dither stacking: If set, common dither positions will not be stacked. This is the default: dither stacking is only recommended for faint spectra that cannot otherwise be automatically extracted.
Ignore dither information from header: If set, all input files are stacked regardless of dither position.
Combination method: Mean is the default; median may also be useful for some input data.
Use weighted mean: If set, the average of the data will be weighted by the variance. Ignored for method=median.
Robust combination: If set, data will be sigma-clipped before combination.
Outlier rejection threshold (sigma): The sigma-clipping threshold for robust combination methods, in units of sigma (standard deviation).
Make Profiles
Wave/space calibration file: The default calibration file is automatically loaded. Set to a valid FITS file path to override the default calibration map with a new one.
Slit correction file: The default slit correction file is automatically loaded, if available. If blank, no slit correction will be applied. Set to a valid FITS file path to override the default file with a new one.
Row fit order: Typically a third-order polynomial fit is used to calculate the smooth spatial profile. Occasionally, a higher or lower order fit may give better results.
Subtract median background: If set, then the median level of the smoothed spatial profile will be subtracted out to remove residual background from the total extracted flux. If the SRCTYPE is EXTENDED_SOURCE this option will be off by default. For other data, this option is appropriate as long as the slit is dominated by background, rather than source flux. If the spatial profile dips below zero at any point (other than for a negative spectrum), this option should be deselected.
Atmospheric transmission threshold: Transmission values below this threshold are not considered when making the spatial profile. Values are 0-1.
Simulate calibrations: Simulate calibration values instead of using the wave/space calibration file. This option is primarily used for testing.
Locate Apertures
Aperture location method: If ‘auto’, the strongest Gaussian peak(s) in the spatial profile will be selected, with an optional starting guess (Aperture position, below). If ‘fix to input’, the value in the Aperture position parameter will be used without refinement. If ‘fix to center’, the center of the slit will be used. ‘Fix to center’ is default for EXTENDED_SOURCE; otherwise ‘auto’ is default.
Number of auto apertures: Set this parameter to 1 to automatically find the single brightest source, or 2 to find the two brightest sources, etc. Sources may be positive or negative.
Aperture position: Enter a guess value for the aperture to use as a starting point for method = ‘auto’, or a fixed value to use as the aperture center for method = ‘fix to input’. Values are in arcseconds up the slit (refer to the spatial profile). Separate multiple apertures for a single file by commas; separate values for multiple files by semi-colons. For example, 3,8;2,7 will look for two apertures in each of two files, near 3” and 8” in the first image and 2” and 7” in the second image. If there are multiple files loaded, but only one aperture list is given, the aperture parameters will be used for all images.
Expected aperture FWHM (arcsec): Gaussian FWHM estimate for spatial profile fits, to determine peaks.
Trace Continuum
Trace method: If ‘fit to continuum’ is selected, points along the continuum will be fit with a Gaussian to determine the trace center at each location, and then the positions will be fit with a low-order polynomial. If ‘fix to aperture position’ is selected, no fit will be attempted, and the default slit curvature defined by the edge definition file will be used as the aperture location. By default, the trace will be fixed for EXTENDED_SOURCE, but a fit will be attempted for all other data types.
Trace fit order: Polynomial fit order for the aperture center, along the spectral dimension.
Trace fit threshold: Sigma value to use for rejecting discrepant trace fits.
Fit position step size (pixels): Step size along the trace for fit positions.
Set Apertures
Extract the full slit: If set, all other parameters are ignored, and the PSF radius will be set to include the full slit.
Refit apertures for FWHM: The spatial FWHM for the aperture is used to determine the aperture and PSF radii, unless they are directly specified. If this parameter is set, the profile will be re-fit with a Gaussian to determine the FWHM. If it is not set, the value determined or set in the Locate Apertures step is used (stored as APFWHM01 in the FITS header).
Aperture sign: Enter either 1 or -1 to skip the automatic determination of the aperture sign from the spatial profile. If the value is -1, the spectrum will be multiplied by -1. Separate multiple apertures by commas; separate values for multiple files by semi-colons. If a single value is specified, it will be applied to all apertures.
Aperture radius: Enter a radius in arcsec to skip the automatic determination of the aperture radius from the profile FWHM. Separate multiple apertures by commas; separate values for multiple files by semi-colons. If a single value is specified, it will be applied to all apertures.
PSF radius: Enter a radius in arcsec to skip the automatic determination of the PSF radius from the profile FWHM. Separate multiple apertures by commas; separate values for multiple files by semi-colons. If a single value is specified, it will be applied to all apertures.
Background regions: Enter a range in arcsec to use as the background region, skipping automatic background determination. For example, 0-1,8-10 will use the regions between 0” and 1” and between 8” and 10” to determine the background level to subtract in extraction. Values are for the full image, rather than for a particular aperture. Separate values for multiple files with semi-colons.
Subtract Background
Skip background subtraction: Set to skip calculating and removing residual background. If no background regions were set, background subtraction will be automatically skipped.
Background fit order: Set to a number greater than or equal to zero for the polynomial order of the fit to the background regions.
Extract Spectra
Save extracted 1D spectra: If set, the extracted spectra will be saved to disk in Spextool format. This option is normally used only for diagnostic purposes.
Extraction method: The default is to use standard extraction for EXTENDED_SOURCE and optimal extraction otherwise. Standard extraction may be necessary for some faint sources.
Use median profile instead of spatial map: By default, the pipeline uses a wavelength-dependent spatial map for extraction, but this method may give poor results if the signal-to-noise in the profile is low. Set this option to use the median spatial profile across all wavelengths instead.
Use spatial profile to fix bad pixels: The pipeline usually uses the spatial profile to attempt to fix bad pixels during standard extraction, and in the 2D image for either extraction method. Occasionally, this results in a failed extraction. Unset this options to extract the spectra without bad pixel correction.
Bad pixel threshold: Enter a value for the threshold for a pixel to be considered a bad pixel. This value is multiplied by the standard deviation of all good pixels in the aperture at each wavelength bin.
Flux Calibrate
General Parameters
Save calibrated 1D spectra: If set, the calibrated spectra will be saved to disk in Spextool format. This option is normally used only for diagnostic purposes.
Skip flux calibration: If set, no telluric correction or flux calibration will be applied.
Response file: The default instrumental response file on disk is automatically loaded. If blank, no response correction will be applied, but transmission correction will still occur. Set to a valid FITS file path to override the default response file with a new one.
Spectral resolution: Expected resolution for the grism mode, used to smooth the ATRAN model. This value should match that of the response file, and should only need modification if the response file is modified from the default.
Telluric Correction Parameters
ATRAN directory: This parameter specifies the location of the library of ATRAN FITS files to use. If blank, the default files provided with the pipeline will be used.
ATRAN file: This parameter is used to override the ATRAN file to use for telluric correction. If blank, the default ATRAN file on disk will be used. Set to a valid FITS file path to override the default ATRAN file with a new one.
Wavelength Shift Parameters
Auto shift wavelength to telluric spectrum: If set, the data will be shifted to match the telluric spectrum. The optimum shift chosen is the one that minimizes residuals in the corrected spectrum, when fit with a low order polynomial. All values within the range of the maximum shift are tested, at a resolution of 0.1 pixels. Auto shift will not be attempted for the FOR_G111 grism.
Maximum auto wavelength shift to apply: The maximum shift allowable for auto-shifts, in pixels.
Wavelength shift to apply: Set to specify a manual shift in pixels along the wavelength axis to apply to the science spectrum. If non-zero, the auto-shift parameter will be ignored.
Polynomial order for continuum: The fit order for the spectrum, used to determine the optimum wavelength shift.
S/N threshold for auto-shift: If the median S/N for a spectrum is below this threshold, auto shift will not be attempted.
Combine Spectra
General Parameters
Registration method: If set to ‘Use WCS as is’, all images will be referenced to the sky position in the first image file. If set to ‘Correct to target position’, the TGTRA/TGTDEC keywords in the FITS header will be used to apply an additional offset for registering non-sidereal targets. If these keywords are not present, or if their value is constant, the algorithm defaults to the ‘Use WCS as is’ behavior. ‘Correct to target position’ is on by default; the other options are recommended only if the TGTRA/TGTDEC or WCS keywords are known to have bad values. In that case, set to ‘Use header offsets’ for non-sidereal targets or files with known bad WCS parameters; otherwise use ‘Use WCS as is’.
Combination method: Mean is the default; median may also be useful for some input data. If ‘spectral_cube’ is set, the input data will be resampled into a 3D spatial/spectral cube instead of coadding 1D spectra and 2D images.
Weight by errors: If set, the average of the data will be weighted by the errors. Ignored for method=median.
1-2D Combination Parameters
Combine apertures: If multiple apertures have been extracted, set this option to combine them into a single 1D spectrum. The 2D image will not attempt to co-align the spectral traces.
Robust combination: If set, data will be sigma-clipped before combination for mean or median methods.
Outlier rejection threshold (sigma): The sigma-clipping threshold for robust combination methods, in units of sigma (standard deviation).
3D Resample Parameters
Spatial surface fit order: This parameter controls the polynomial order of the surface fit to the data at each grid point. Higher orders give more fine-scale detail, but are more likely to be unstable. Set to zero to do a weighted mean of the nearby data.
Spatial fit window: Spatial window (pixels) for consideration in local data fits. Set higher to fit to more pixels.
Spatial smoothing radius: Gaussian width (pixels) for smoothing radius in distance weights for local data fits. Set higher to smooth over more pixels.
Spatial edge threshold: A value between 0 and 1 that determines how much of the image edge is set to NaN. Set higher to set more pixels to NaN.
Adaptive smoothing algorithm: If ‘scaled’, the size of the smoothing kernel is allowed to vary, in order to optimize reconstruction of sharply peaked sources. If ‘shaped’, the kernel shape and rotation may also vary. If ‘none’, the kernel will not vary.
Make Spectral Map
Color map: Color map for the output PNG image. Any valid Matplotlib name may be specified.
Flux scale for image: A low and high percentile value , used for scaling the spectral image, e.g. [0,99].
Number of contours: Number of contour levels to be over-plotted on the image. Set to 0 to turn off contours.
Contour color: Color for the contour lines. Any valid Matplotlib color name may be specified.
Filled contours: If set, contours will be filled instead of overlaid.
Overlay grid: If set, a coordinate grid will be overlaid.
Watermark text: If set to a non-empty string, the text will be added to the lower-right of the image as a semi-transparent watermark.
Fraction of outer wavelengths to ignore: Used to block edge effects for noisy spectral orders. Set to 0 to include all wavelengths in the plot.
Overplot transmission: If set, the atmospheric transmission spectrum will be displayed in the spectral plot.
Flux scale for spectral plot: Specify a low and high percentile value for the spectral flux scale, e.g. [0,99]. If set to [0, 100], Matplotlib defaults are used.
Make Response
Standard model file: If blank, a model file will be searched for in the default data directory. Set to a valid FITS file to override.
Data quality assessment¶
After the pipeline has been run on a set of input data, the output products should be checked to ensure that the data has been properly reduced. Data quality and quirks can vary widely across individual observations, but the following general guideline gives some strategies for approaching quality assessment (QA) for FLITECAM data.
Check for QA comments in the FITS header HISTORY. These comments may make suggestions for files to exclude from final reductions, or for non-default parameters to set for optimal reductions.
Check the output to the log file (usually called redux_[date]_[time].log), written to the same directory as the output files. Look for messages marked ERROR or WARNING. The log will also list every parameter used in the pipeline steps, which may help disambiguate the parameters as actually-run for the pipeline.
Check that the expected files were written to disk: there should, at a minimum, be a calibrated file (CAL) for imaging data and a combined spectrum (CMB) for grism data. Check the data product tables (Table 9 and Table 10) for other expected data products for each mode.
For imaging data:
Check the flat frame in the FLT file by comparing it with the raw image. All instrumental artifacts (areas of low quantum efficiency, obscurations in the optical path, and other systematics) that are present in the raw frame should be present in the flat. Sources in the raw image should not appear in the flat. Also check that the gain-corrected image does not contain any residual artifacts.
Check for excessive hot or cold pixels in the coadded image. Bad pixels should be ignored in the coadding process.
Check that the background was correctly subtracted. The counts in regions containing no sources should be zero, within the standard deviation.
Check that the registration process calculated offsets correctly: compare all the registered images to verify that all sources appear at the same location in the WCS.
For grism data:
Display the spatial profile with apertures overlaid. Verify that apertures look well placed and the spatial profile does not dip below zero (except for negative spectral traces).
Display the rectified image, and overlay the locations of the extracted apertures. Verify that the apertures lie on top of any visible spectral traces.
Display the intermediate spectra, and verify that all spectra of the same target look similar.
Display the final spectrum (CMB) and overplot the expected atmospheric transmission. Check that the calibrated spectrum does not include residual artifacts from the telluric absorption features. If it does, the assumed resolution for the grism, or the wavelength calibration of the observation, may need updating.
Overlay a model spectrum on the calibrated spectra of flux standards. Verify that the observed spectrum matches the theoretical spectrum, within the error bars of the observation. If it does not, the instrumental response file may need updating.
Appendix A: Sample configuration files¶
Below are sample FLITECAM Redux parameter override files in INI format. If present, the parameter value overrides the default defined by the FLITECAM reduction object. If not present, the default value will be used.
# Redux parameters for FLITECAM instrument in imaging mode
# Pipeline: FLITECAM_REDUX v2_0_0
[1: check_header]
abort = True
[2: correct_linearity]
save = False
linfile = linearity_files/lc_coeffs_20140325.fits
saturation = 5000
[3: clip_image]
save = False
skip_clean = False
datasec = 186, 838, 186, 838
[4: make_flat]
save = True
flatfile = ""
skip_flat = False
[5: correct_gain]
save = False
[6: subtract_sky]
save = False
skyfile = ""
skip_sky = False
sky_method = Use image median
[7: register]
save = False
corcoadd = Use WCS as is
offsets = ""
mfwhm = 6
xyshift = 100
[8: tellcor]
save = True
[9: coadd]
save = True
skip_coadd = False
reference = Target position
method = median
weighted = True
robust = True
threshold = 8.0
maxiters = 5
smoothing = 1.0
[10: fluxcal]
save = True
rerun_phot = False
srcpos = ""
fitsize = 138
fwhm = 6.0
profile = Moffat
[11: imgmap]
colormap = plasma
scale = 0.25, 99.9
n_contour = 0
contour_color = gray
fill_contours = False
grid = True
beam = False
watermark = ""
# Redux parameters for FLITECAM instrument in spectroscopy mode
# Pipeline: FLITECAM_REDUX v2_0_0
[1: check_header]
abort = True
[2: correct_linearity]
save = False
linfile = linearity_files/lc_coeffs_20140325.fits
saturation = 5000
[3: make_image]
save = True
pair_sub = True
flatfile = grism/Cals_20151006/Flats/FLT_A1_LM_flat.fits
[4: stack_dithers]
save = True
skip_stack = True
ignore_dither = False
method = mean
weighted = True
robust = True
threshold = 8.0
maxiters = 5
[5: make_profiles]
save = True
wavefile = grism/Cals_20151006/2dWaveCals/flt_a1_lm_map2pos_ngc7027.fits
slitfile = ""
fit_order = 3
bg_sub = True
atmosthresh = 0.0
simwavecal = False
[6: locate_apertures]
save = False
method = auto
num_aps = 2
input_position = ""
fwhm = 3.0
[7: trace_continuum]
save = False
method = fit to continuum
fit_order = 2
fit_thresh = 4.0
step_size = 9
[8: set_apertures]
save = False
full_slit = False
refit = True
apsign = ""
aprad = ""
psfrad = ""
bgr = ""
[9: subtract_background]
save = False
skip_bg = False
bg_fit_order = 0
[10: extract_spectra]
save = True
save_1d = True
method = optimal
use_profile = False
fix_bad = True
threshold = 4.0
[11: flux_calibrate]
save = True
save_1d = False
skip_cal = False
respfile = grism/response/v4.0.0/FC_GRI_A1LM_SS20_RSP.fits
resolution = 1075.0
atrandir = ""
atranfile = ""
auto_shift = True
auto_shift_limit = 2.0
waveshift = 0.0
model_order = 1
sn_threshold = 10.0
[12: combine_spectra]
save = True
registration = Correct to target position
method = mean
weighted = True
combine_aps = True
robust = True
threshold = 8.0
maxiters = 5
fit_order = 2
fit_window = 7.0
smoothing = 2.0
edge_threshold = 0.7
adaptive_algorithm = none
[13: specmap]
colormap = plasma
scale = 0.25, 99.9
n_contour = 0
contour_color = gray
fill_contours = False
grid = False
watermark = ""
ignore_outer = 0.0
atran_plot = True
spec_scale = 0.25, 99.75
Appendix B: Required input keywords¶
The files below define all keywords that the FLITECAM pipeline checks for validity before proceeding. They are normally located in the pipeline distribution distribution at sofia_redux/instruments/flitecam/data/keyword_files.
# FLITECAM imaging header requirements configuration file
#
# Keywords in this list are only those required for successful
# data reduction (grouping and processing). There may be more
# keywords required by the SOFIA DCS.
#
# Requirement value should be *, nodding, or dithering,
# (as denoted by the corresponding FITS keywords).
# * indicates a keyword that is required for all data. All
# others will only be checked if they are appropriate to the
# mode of the input data.
#
# DRange is not required to be present in the configuration --
# if missing, the keyword will be checked for presence only. If
# drange is present, it will be checked for an enum requirement
# first; other requirements are ignored if present. Min/max
# requirements are only used for numerical types, and are inclusive
# (i.e. the value may be >= min and <= max).
#
# 2021-02-19 Melanie Clarke: First version
[ALTI_STA]
requirement = *
dtype = float
[[drange]]
min = 0
max = 60000
[COADDS]
requirement = *
dtype = int
[[drange]]
min = 1
[CYCLES]
requirement = *
dtype = int
[[drange]]
min = 0
[DATE-OBS]
requirement = *
dtype = str
[DITHER]
requirement = *
dtype = bool
[DIVISOR]
requirement = *
dtype = int
[[drange]]
min = 1
[EXPTIME]
requirement = *
dtype = float
[[drange]]
min = 0.0
[INSTCFG]
requirement = *
dtype = str
[[drange]]
enum = IMAGING
[INSTMODE]
requirement = *
dtype = str
[[drange]]
enum = STARE, NOD_OFFARRAY
[INSTRUME]
requirement = *
dtype = str
[[drange]]
enum = FLITECAM
[ITIME]
requirement = *
dtype = float
[[drange]]
min = 0.0
[MISSN-ID]
requirement = *
dtype = str
[NDR]
requirement = *
dtype = int
[[drange]]
min = 1
max = 32
[NODDING]
requirement = *
dtype = bool
[NODBEAM]
requirement = nodding
dtype = str
[[drange]]
enum = A, B
[OBJECT]
requirement = *
dtype = str
[OBS_ID]
requirement = *
dtype = str
[OBSTYPE]
requirement = *
dtype = str
[[drange]]
enum = OBJECT, STANDARD_FLUX, FLAT, SKY
[SPECTEL1]
requirement = *
dtype = str
[[drange]]
enum = FLT_J, FLT_H, FLT_K, FLT_ICE_308, FLT_PAH_329, FLT_Pa, FLT_Pa_cont, FLT_NbL, FLT_NbM, FLT_L, FLT_Lprime, FLT_M
[SPECTEL2]
requirement = *
dtype = str
[[drange]]
enum = NONE
[SRCTYPE]
requirement = *
dtype = str
[[drange]]
enum = POINT_SOURCE, COMPACT_SOURCE, EXTENDED_SOURCE, OTHER, UNKNOWN
[TABLE_MS]
requirement = *
dtype = float
[[drange]]
min = 0
[ZA_START]
requirement = *
dtype = float
[[drange]]
min = 0
max = 90
# FLITECAM grism header requirements configuration file
#
# Keywords in this list are only those required for successful
# data reduction (grouping and processing). There may be more
# keywords required by the SOFIA DCS.
#
# Requirement value should be *, nodding, or dithering,
# (as denoted by the corresponding FITS keywords).
# * indicates a keyword that is required for all data. All
# others will only be checked if they are appropriate to the
# mode of the input data.
#
# DRange is not required to be present in the configuration --
# if missing, the keyword will be checked for presence only. If
# drange is present, it will be checked for an enum requirement
# first; other requirements are ignored if present. Min/max
# requirements are only used for numerical types, and are inclusive
# (i.e. the value may be >= min and <= max).
#
# 2021-02-19 Melanie Clarke: First version
[ALTI_STA]
requirement = *
dtype = float
[[drange]]
min = 0
max = 60000
[COADDS]
requirement = *
dtype = int
[[drange]]
min = 1
[CYCLES]
requirement = *
dtype = int
[[drange]]
min = 0
[DATE-OBS]
requirement = *
dtype = str
[DITHER]
requirement = *
dtype = bool
[DIVISOR]
requirement = *
dtype = int
[[drange]]
min = 1
[EXPTIME]
requirement = *
dtype = float
[[drange]]
min = 0.0
[INSTCFG]
requirement = *
dtype = str
[[drange]]
enum = SPECTROSCOPY, GRISM
[INSTMODE]
requirement = *
dtype = str
[[drange]]
enum = STARE, NOD_ALONG_SLIT, NOD_OFF_SLIT
[INSTRUME]
requirement = *
dtype = str
[[drange]]
enum = FLITECAM
[ITIME]
requirement = *
dtype = float
[[drange]]
min = 0.0
[MISSN-ID]
requirement = *
dtype = str
[NDR]
requirement = *
dtype = int
[[drange]]
min = 1
max = 32
[NODDING]
requirement = *
dtype = bool
[NODBEAM]
requirement = nodding
dtype = str
[[drange]]
enum = A, B
[OBJECT]
requirement = *
dtype = str
[OBS_ID]
requirement = *
dtype = str
[OBSTYPE]
requirement = *
dtype = str
[[drange]]
enum = OBJECT, STANDARD_TELLURIC, FLAT, SKY
[SLIT]
requirement = *
dtype = str
[[drange]]
enum = FLT_SS10, FLT_SS20
[SPECTEL1]
requirement = *
dtype = str
[[drange]]
enum = FLT_A1_LM, FLT_A2_KL, FLT_A3_Hw, FLT_B1_LM, FLT_B2_Hw, FLT_B3_J, FLT_C2_LM, FLT_C3_Kw, FLT_C4_H
[SPECTEL2]
requirement = *
dtype = str
[[drange]]
enum = FLT_SS10, FLT_SS20
[SRCTYPE]
requirement = *
dtype = str
[[drange]]
enum = POINT_SOURCE, COMPACT_SOURCE, EXTENDED_SOURCE, OTHER, UNKNOWN
[TABLE_MS]
requirement = *
dtype = float
[[drange]]
min = 0
[ZA_START]
requirement = *
dtype = float
[[drange]]
min = 0
max = 90
Appendix C: Calibration Data Generation¶
The FLITECAM Redux pipeline requires several kinds of auxiliary reference calibration files, listed in Table 13. Some of these are produced by tools packaged with the pipeline. This section describes the procedures used to produce these auxiliary files.
Instrumental Response Curve¶
As described above, instrumental response curves are automatically produced for each spectrum with OBSTYPE = STANDARD_TELLURIC. For use in calibrating science spectra, response curves from multiple observations must be combined together.
For appropriate combination, input response curves must share the same grism, slit, and detector bias setting.
Matching response curves may be scaled, to account for variations in slit loss or model accuracy, then are generally combined together with a robust weighted mean statistic. The combined curve is smoothed with a Gaussian of width 2 pixels, to reduce artificial artifacts. Averaged response curves for each grism and slit combination are usually produced for each flight series, and stored for pipeline use in the standard location for the instrument package (data/grism/response).
The scaling, combination, and smoothing of instrument response curves is implemented as a final step in the pipeline for grism standards. After individual response_spectrum files (*RSP*.fits) are grouped appropriately, the final step in the pipeline can be run on each group to produce the average instrument_response file (*IRS*.fits).
Useful Parameters¶
Below are some useful parameters for combining response spectra.
Combine Response
Scaling Parameters
Scaling method: If ‘median’, all spectra are scaled to the median of the flux stack. If ‘highest’, all spectra are scaled to the spectrum with the highest median value. If ‘lowest’, all spectra are scaled to the spectrum with the lowest median value. If ‘index’, all spectra are scaled to the spectrum indicated in the Index parameter, below. If ‘none’, no scaling is applied before combination.
Index of spectrum to scale to: If Scaling method is ‘index’, set this value to the index of the spectrum to scale. Indices start at zero and refer to the position in the input file list.
Combination Parameters
Combine apertures: For multi-aperture data, it may be useful to produce a separate response curve for each aperture. Select this option to combine them into a single reponse curve instead.
Combination method: Mean is default; median may also be useful for some input data.
Weight by errors: If set, the average of the data will be weighted by the variance. Ignored for method=median.
Robust combination: If set, data will be sigma-clipped before combination for mean or median methods.
Outlier rejection threshold (sigma): The sigma-clipping threshold for robust combination methods, in units of sigma (standard deviation).
Smoothing Parameters
Smoothing Gaussian FWHM: Full-width-half-max for the Gaussian kernel used for smoothing the final response spectrum, specified in pixels.
Wavelength Calibration Map¶
Calibration Principles¶
Grism wavelength and spatial calibrations are stored together in a single image extension in a FITS file, where the first plane is the wavelength calibration and the second is the spatial calibration. The images should match the dimensions of the raw data arrays, assigning a wavelength value in um and a slit position in arcsec to every raw pixel.
These calibration files are generally derived from specialized calibration data. Wavelength calibration is best derived from images for which strong emission or absorption lines fill the whole image, from top to bottom, and evenly spaced from left to right. Sky data may be used for this purpose for some of the grism passbands; lab data may be more appropriate for others. Raw data should be cleaned and averaged or summed to produce an image with as high a signal-to-noise ratio in the spectroscopic lines as possible.
After preprocessing, the spectroscopic lines must be identified with specific wavelengths from a priori knowledge, then they must be re-identified with a centroiding algorithm at as many places across the array as possible. The identified positions can then be fit with a smooth 2D surface, which provides the wavelength value in microns at any pixel, accounting for any optical distortions as needed.
In principle, the spatial calibration proceeds similarly. Spatial calibrations are best derived from identifiable spectral continuua that fill the whole array from left to right, evenly spaced from top to bottom. Most commonly, special observations of a spectroscopic standard are taken, placing the source at multiple locations in the slit. These spectroscopic traces are identified then re-fit across the array. The identified positions are again fit with a smooth 2D surface to provide the spatial position in arcseconds up the slit at any pixel. This calibration can then be used to correct for spatial distortions, in the same way that the wavelength calibration is used to rectify distortions along the wavelength axis.
Pipeline Interface¶
The input data for calibration tasks is generally raw FITS files, containing spectroscopic data. In order to perform calibration steps instead of the standard spectroscopic pipeline, the pipeline interface requires a user-provided flag, either in an input configuration file, or on the command line, as for example:
redux_pipe -c wavecal=True /path/to/fits/files
for a wavelength calibration reduction or:
redux_pipe -c spatcal=True /path/to/fits/files
for a spatial calibration reduction.
The first steps in either reduction mode are the same pre-processing steps used in the standard pipeline reduction. The stacking steps have optional parameters that allow for the input data to be summed instead of subtracted (for calibration from sky lines), or to be summed instead of averaged (for combining multiple spectral traces into a single image).
Thereafter, the wavecal reduction performs the following steps. Each step has a number of tunable parameters; see below for parameter descriptions.
Make Profiles: a spatial profile is generated from the unrectified input image.
Extract First Spectrum: an initial spectrum is extracted from a single aperture, via a simple sum over a specified number of rows.
Identify Lines: spectrosopic lines specified in an input list are identified in the extracted spectrum, via Gaussian fits near guess positions derived from user input or previous wavelength calibrations.
Reidentify Lines: new spectra are extracted from the image at locations across the array, and lines successfully identified in the initial spectrum are attempted to be re-identified in each new spectrum.
Fit Lines: all input line positions and their assumed wavelength values are fit with a low-order polynomial surface. The fit surface is saved to disk as the wavelength calibration file.
Verify Rectification: the derived wavelength calibration is applied to the input image, to verify that correctly rectifies the spectral image.
After preprocessing, the spatcal reduction performs similar steps:
Make Profiles: a spatial profile is generated from the unrectified input image.
Locate Apertures: spectral apertures are identified from the spatial profile, either manually or automatically.
Trace Continuum: spectrosopic continuuum positions are fit in steps across the array, for each identified aperture.
Fit Traces: all aperture trace positions are fit with a low-order polynomial surface. The fit surface is saved to disk as the spatial calibration file.
Verify Rectification: the derived spatial calibration is applied to the input image, to verify that correctly rectifies the spectral image.
Intermediate data can also be saved after any of these steps, and can be later loaded and used as a starting point for subsequent steps, just as in the standard spectroscopic pipeline. Parameter settings can also be saved in a configuration file, for later re-use or batch processing.
Wavelength and spatial calibrations generally require different pre-processing steps, or different input data altogether, so they cannot be generated at the same time. The pipeline interface will allow a previously generated wavelength or spatial calibration file to be combined together with the new one in the final input. Optional previous spatial calibration input is provided to the wavecal process in the Fit Lines step; optional previous wavelength calibration input is provided to the spatcal process in the Fit Traces step. If a previously generated file is not provided, the output file will contain simulated data in the spatial or wavelength plane, as appropriate.
Reference Data¶
Line lists for wavelength calibration are stored in the standard reference data directory for the instrument package (data/grism/line_lists). In these lists, commented lines (beginning with ‘#’) are used for display only; uncommented lines are attempted to be fit. Initial guesses for the pixel position of the line may be taken from a previous wavelength calibration, or from a low-order fit to wavelength/position pairs input by the user. Default wavelength calibration files and line lists may be set by date, in the usual way (see data/grism/caldefault.txt).
Spatial calibration uses only the assumed slit height in pixels and arcsec as input data, as stored in the reference files in data/grism/order_mask. These values are not expected to change over time.
Display Tools¶
The pipeline incorporates several display tools for diagnostic purposes. In addition to the DS9 display of the input and intermediate FITS files, spatial profiles and extracted spectra are displayed in separate windows, as in the standard spectroscopic pipeline. Identified lines for wavecal are marked in the spectral display window (Fig. 63); identified apertures for spatcal are marked in the spatial profile window (Fig. 64). Fit positions and lines of constant wavelength or spatial position are displayed as DS9 regions. These region files are also saved to disk, for later analysis. Finally, after the line or trace positions have been fit, a plot of the residuals, against X and Y position is displayed in a separate window (Fig. 65 and Fig. 66). This plot is also saved to disk, as a PNG file.
Useful Parameters¶
Some key parameters used specifically for the calibration modes are listed below. See above for descriptions of parameters for the steps shared with the standard pipeline.
Wavecal Mode¶
Stack Dithers
Ignore dither information from header: This option allows all input dithers to be combined together, regardless of the dither information in the header. This option may be useful in generating a high signal-to-noise image for wavelength identification.
Extract First Spectrum
Save extracted 1D spectra: If set, a 1D spectrum is saved to disk in Spextool format. This may be useful for identifying line locations in external interactive tools like xvspec (in the IDL Spextool package).
Aperture location method: If ‘auto’, the most significant peak in the spatial profile is selected as the initial spectrum region, and the aperture radius is determined from the FWHM of the peak. If ‘fix to center’, the center pixel of the slit is used as the aperture location. If ‘fix to input’, the value specified as the aperture position is used as the aperture location.
Polynomial order for spectrum detrend: If set to an integer 0 or higher, the extracted spectrum will be fit with a low order polynomial, and this fit will be subtracted from the spectrum. This option may be useful to flatten a spectrum with a a strong trend, which can otherwise interfere with line fits.
Identify Lines
Wave/space calibration file: A previously generated wavelength calibration file, to use for generating initial guesses of line positions. If a significant shift is expected from the last wavelength calibration, the ‘Guess’ parameters below should be used instead.
Line list: List of wavelengths to fit in the extracted spectrum. Wavelengths should be listed, one per line, in microns. If commented out with a ‘#’, the line will be displayed in the spectrum as a dotted line, but a fit to it will not be attempted.
Line type: If ‘absorption’, only concave lines will be expected. If ‘emission’, only convex lines are expected. If ‘either’, concave and convex lines may be fit. Fit results for faint lines are generally better if either ‘absorption’ or ‘emission’ can be specified.
Fit window: Window (in pixels) around the guess position used as the fitting data. Smaller windows may result in more robust fits for faint lines, if the guess positions are sufficiently accurate.
Expected line width (pixel): FWHM expected for the fit lines.
Guess wavelengths: Comma-separated list of wavelengths for known lines in the extracted spectrum. If specified, must match the list provided for Guess wavelength position, and the Wave/space calibration file will be ignored. If two values are provided, they will be fit with a first-order polynomial to provide wavelength position guesses for fitting. Three or more values will be fit with a second-order polynomial.
Guess wavelength position: Comma-separated list of pixel positions for known lines in the image. Must match the provided Guess wavelengths.
Reidentify Lines
Save extracted 1D spectra: If set, all extracted spectra are saved to disk in Spextool format, for more detailed inspection and analysis.
Aperture location method: If ‘step up slit’, apertures will be placed at regular intervals up the slit, with step size specified in Step size and radius specified in Aperture radius. If ‘fix to input’, then apertures will be at the locations specified by Aperture position and radius specified in Aperture radius. If ‘auto’, apertures will be automatically determined from the spatial profile.
Number of auto apertures: If Aperture location method is ‘auto’, this many apertures will be automatically located.
Aperture position: Comma-separated list of aperture positions in pixels. Apertures in multiple input files may also be specified, using semi-colons to separate file input. If Aperture location method is ‘auto’, these will be used as starting points. If ‘fix to input’, they will be used directly.
Aperture radius: Width of the extracted aperture, in pixels. The radius may be larger than the step, allowing for overlapping spectra. This may help get higher S/N for extracted spectra in sky frames.
Polynomial order for spectrum detrend: As for the Extract First Spectrum step, setting this parameter to an integer 0 or higher will detrend it. If detrending is used for the earlier step, it is recommended for this one as well.
Fit window: Window (in pixels) around the guess position used as the fitting data. The guess position used is the position in the initial spectrum, so this window must be wide enough to allow for any curvature in the line.
Signal-to-noise requirement: Spectral S/N value in sigma, below which a fit will not be attempted at that line position in that extracted spectrum.
Fit Lines
Fit order for X: Polynomial surface fit order in the X direction. Orders 2-4 are recommended.
Fit order for Y: Polynomial surface fit order in the Y direction. Orders 2-4 are recommended.
Weight by line height: If set, the surface fit will be weighted by the height of the line at the fit position. This can be useful if there is a good mix of strong and weak lines across the array. If there is an imbalance of strong and weak lines across the array, this option may throw the fit off at the edges.
Spatial calibration file: If provided, the spatial calibration plane in the specified file will be combined with the wavelength fit to produce the output calibration file (*WCL*.fits). The default is the wavelength calibration file from the previous series. If not provided, a simulated flat spatial calibration will be produced and attached to the output calibration file.
Spatcal Mode¶
Aperture location and continuum tracing follow the standard spectroscopic method, with the exception that units are all in pixels rather than arcseconds. See above for descriptions of the parameters for the Locate Apertures and Trace Continuum steps.
See the wavecal mode descriptions, above, for useful parameters for the Stack and Stack Dithers steps.
Fit Trace Positions
Fit order for X: Polynomial surface fit order in the X direction. Orders 2-4 are recommended.
Fit order for Y: Polynomial surface fit order in the Y direction. Orders 2-4 are recommended.
Weight by profile height: If set, the surface fit will be weighted by the height of the aperture in the spatial map at the fit position.
Wavelength calibration file: If provided, the wavelength calibration plane in the specified file will be combined with the spatial fit to produce the output calibration file (*SCL*.fits). The default is the wavelength calibration file from the previous series. If not provided, pixel positions will be stored in the wavelength calibration plane in the output file.
Slit Correction Image¶
The response spectra used to flux-calibrate spectroscopic data encode variations in instrument response in the spectral dimension, but do not account for variations in response in the spatial dimension. For compact sources, spatial response variations have minimal impact on the extracted 1D spectrum, but for extended targets or SLITSCAN observations, they should be corrected for.
To do so, the pipeline divides out a flat field, called a slit correction image, that contains normalized variations in response in the spatial dimension only.
These slit correction images can be derived from wavelength-rectified sky frames, as follows:
Median spectra are extracted at regular positions across the frame.
All spectra are divided by the spectrum nearest the center of the slit.
The normalized spectra are fit with a low-order polynomial to derive smooth average response variations across the full array.
The fit surface is the slit correction image. It is stored as a single extension FITS image, and can be provided to the standard spectroscopic pipeline at the Make Profiles step. These images should be regenerated whenever the wavelength and spatial calibrations are updated, since the slit correction image matches the rectified dimensions of the spectral data, not the raw dimensions.
Pipeline Interface¶
Similar to the wavecal and spatcal modes described above, the pipeline provides a slitcorr mode to produce slit correction images starting from raw FITS files. This mode can be invoked with a configuration flag:
redux_pipe -c slitcorr=True /path/to/fits/files
The pre-processing steps in slitcorr reduction mode are the same as in the standard pipeline reduction, except that the default for the stacking steps is to add all chop/nod frames and average all input files, to produce a high-quality sky frame. Rectification and spatial profile generation also proceeds as usual, using the latest available wavelength calibration file.
Thereafter, the slitcorr reduction performs the following steps. Each step has a number of tunable parameters; see below for parameter descriptions.
Locate Apertures: a number of apertures are spaced evenly across the slit.
Extract Median Spectra: flux data is median-combined at each wavelength position for each aperture.
Normalize Response: median spectra are divided by the spectrum nearest the center of the slit. The 2D flux image is similarly normalized, for reference.
Make Slit Correction: the normalized spectra are fit with a low-order polynomial to produce a smooth slit correction surface that matches the rectified data dimensions.
Intermediate data can also be saved after any of these steps, and can be later loaded and used as a starting point for subsequent steps, just as in the standard spectroscopic pipeline. Parameter settings can also be saved in a configuration file, for later re-use or batch processing.
Useful Parameters¶
Some key parameters used specifically for the slitcorr mode are listed below. See above for descriptions of parameters for the steps shared with the standard pipeline.
Locate Apertures
Number of apertures: For this mode, apertures are evenly spaced across the array. Specify the desired number of apertures. The radius for each aperture is automatically assigned to not overlap with its neighbors.
Extract Median Spectra
Save extracted 1D spectra: If set, all extracted spectra are saved to disk in a FITS file in Spextool format, for inspection.
Normalize Response
Save extracted 1D spectra: Save normalized spectra to disk in Spextool format.
Make Slit Correction
General Parameters
Fit method: If ‘2D’, a single surface is fit to all the normalized spectral data, producing a smooth low-order polynomial surface. If ‘1D’, polynomial fits are performed in the y-direction only, at each wavelength position, then are smoothed in the x-direction with a uniform (boxcar) filter. The 1D option may preserve higher-order response variations in the x-direction; the 2D option will produce a smoother surface.
Weight by spectral error: If set, the polynomial fits will be weighted by the error propagated for the normalized median spectra.
Parameters for 2D fit
Fit order for X: Polynomial surface fit order in the X direction. Orders 2-4 are recommended.
Fit order for Y: Polynomial surface fit order in the Y direction. Orders 2-4 are recommended.
Parameters for 1D fit
Fit order for Y: Polynomial fit order in the Y direction. Orders 2-4 are recommended.
Smoothing window for X: Boxcar width for smoothing in X direction, in pixels.
Fig. 63 Wavecal mode reduction and diagnostic plots.¶
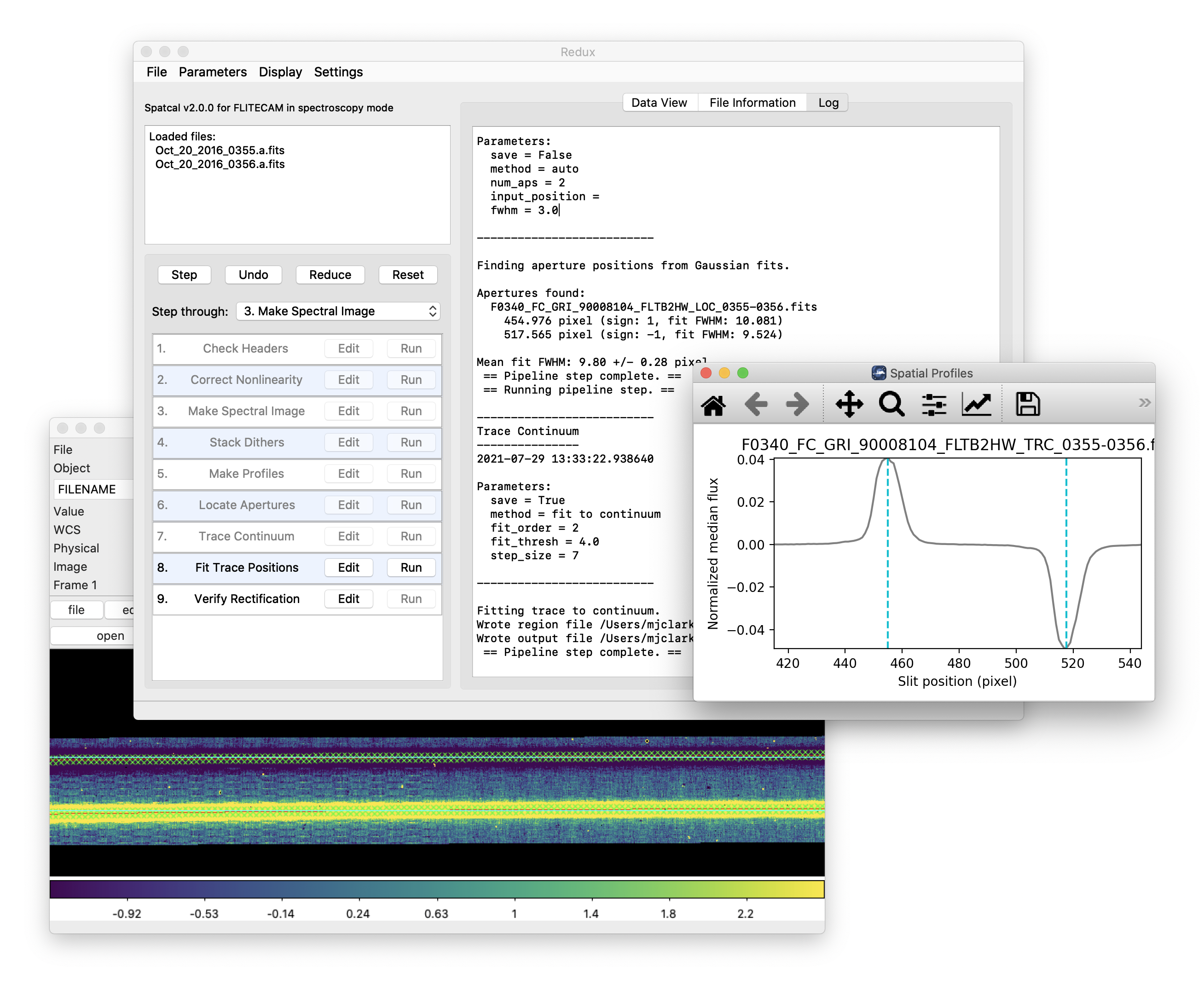
Fig. 64 Spatcal mode reduction and diagnostic plots.¶
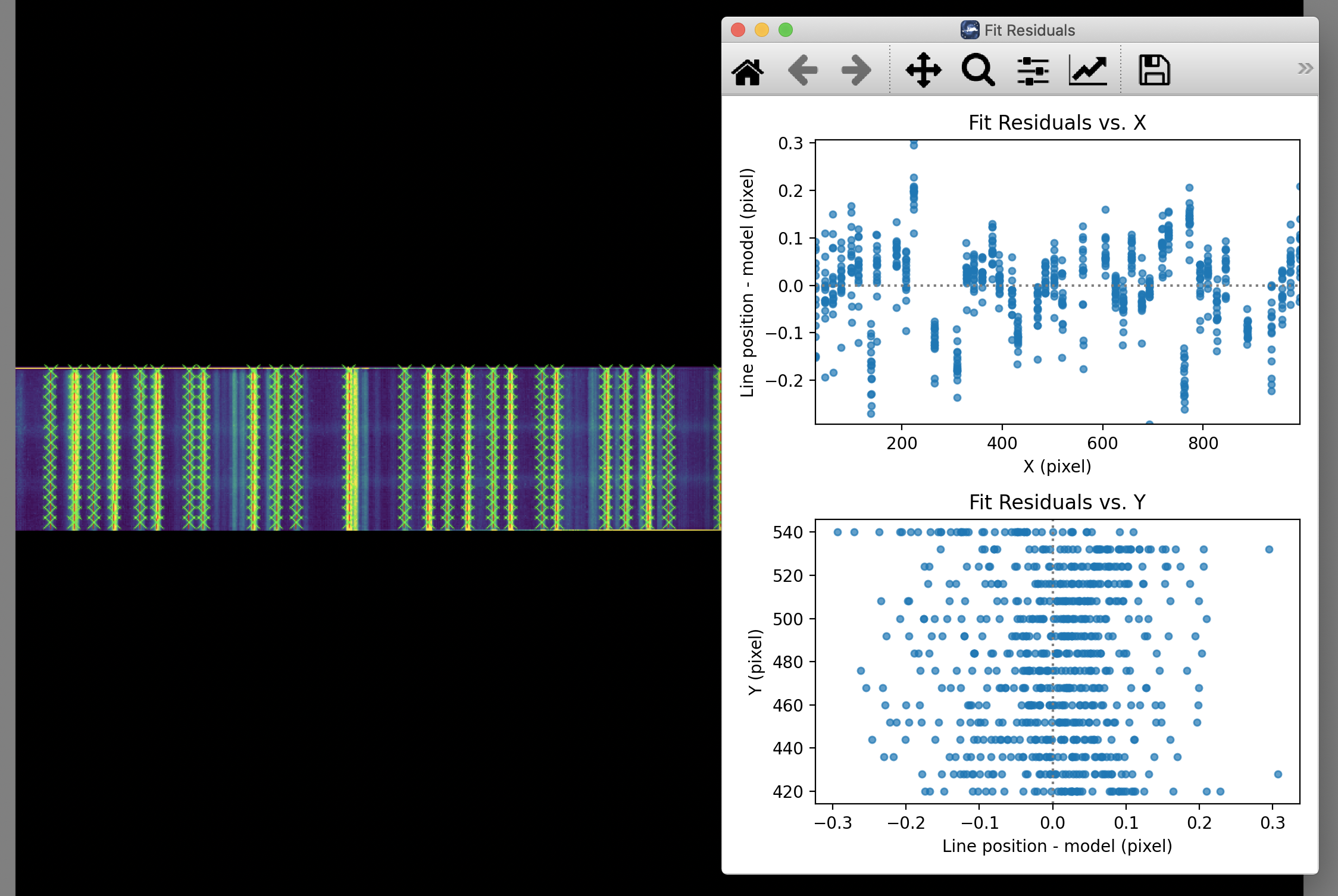
Fig. 65 Wavecal mode fit surface and residuals.¶
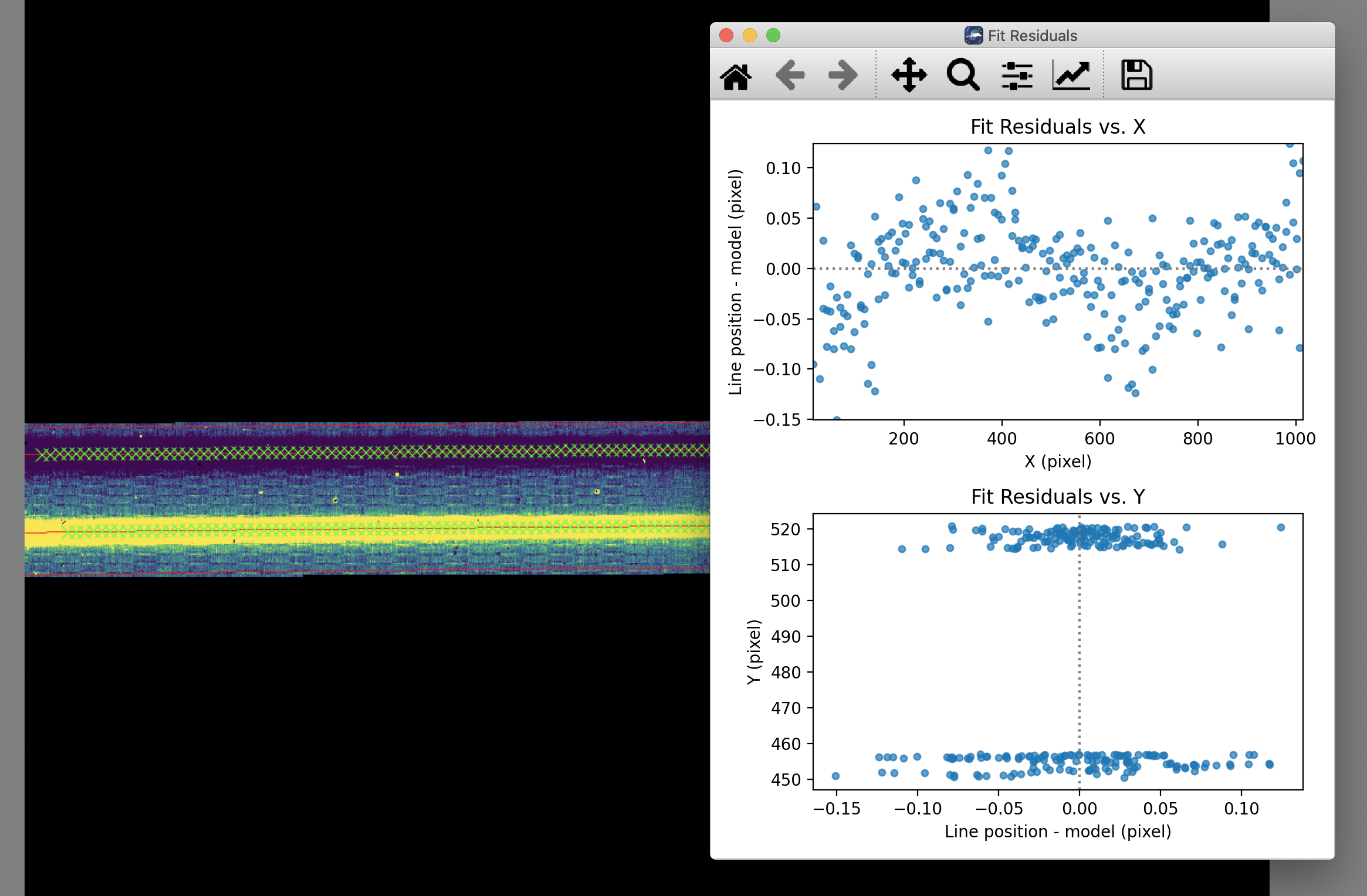
Fig. 66 Spatcal mode fit surface and residuals.¶
Appendix D: Change notes for the FLITECAM pipeline¶
Significant changes¶
Below are listed the most significant changes for the FLITECAM pipeline over its history, highlighting impacts to science data products. See the data handbooks or user manuals associated with each release for more information.
All pipeline versions prior to v2.0.0 were implemented in IDL; v2.0.0 and later were implemented in Python. An early predecessor to the FLITECAM Redux pipeline, called FDRP/FSpextool, was also released for FLITECAM reductions in 2013, but no data in the SOFIA archive remains that was processed with this pipeline.
For previously processed data, check the PIPEVERS keyword in the FITS header to determine the pipeline version used.
FLITECAM Redux v2.0.0 (2021-09-24)¶
User manual: Rev. B
All modes¶
Full reimplementation of the IDL pipeline into Python 3.
Imaging¶
Data formats change significantly. Imaging products now separate flux, error, and exposure map into separate FITS image extensions, rather than storing them as a 3D cube in the primary extension.
Spectroscopy¶
Data formats change significantly. Images and spectra are stored in the same FITS file, under separate extensions. Final 1D spectra (CMB files, PRODTYPE=combined_spectrum) are still stored in the same format as before; the spectrum corresponds to the SPECTRAL_FLUX extension in the COA (PRODTYPE=coadded_spectrum) file.
FLITECAM Redux v1.2.0 (2017-12-15)¶
User manual: Rev. A
Imaging¶
Flux calibration procedure revised to separate telluric correction from flux calibration. Telluric correction is now performed on a file-by-file basis, for better accuracy, after registration. The REG file is no longer saved by default; it is replaced by a TEL file which is telluric-corrected but not flux calibration. The final calibration factor is still applied at the end of the pipeline, making a single CAL file. The CALFCTR stored in the header is now the calibration factor at the reference altitude and zenith angle; it no longer includes the telluric correction factor. The latter value is stored in the new keyword TELCORR.
Image registration default set to use the WCS for most image shifts, instead of centroid or cross-correlation algorithms.
FLITECAM Redux v1.1.0 (2016-09-20)¶
User manual: Rev. A
Imaging¶
Flux calibration factors are now applied to data arrays to convert them to physical units (Jy). The calibrated data product has file code CAL (PRODTYPE=calibrated). COA files are no longer designated Level 3, even if their headers contain calibration factors.
Spectroscopy¶
Grism calibration incorporated into the pipeline, using stored instrumental response files, similar to the FORCAST grism calibration process.
FLITECAM Redux v1.0.3 (2015-10-06)¶
User manual: Rev. -
All modes¶
Minor bug fixes for filename handling and batch processing.
FLITECAM Redux v1.0.2 (2015-09-03)¶
User manual: Rev. -
Imaging¶
Improvements to the flat generation procedures.
FLITECAM Redux v1.0.1 (2015-05-14)¶
User manual: Rev. -
All modes¶
EXPTIME keyword updated to track total nominal on-source integration time.
ASSC_AOR keyword added to track all input AOR-IDs.
Imaging¶
Separate flat and sky files accommodated.
Flux calibration incorporated into pipeline, rather than applied as a separate step.
FLITECAM Redux v1.0.0 (2015-01-23)¶
User manual: Rev. -
All modes¶
Integrated FLITECAM imaging algorithms (FDRP) with Spextool spectral extraction algorithms, in a standard pipeline interface (Redux).